There has been quite a lot of chatter recently about different scientific publishing models. Prompted by Elsevier’s support for the Research Works Act, and the resulting proposed academic boycott.
Let there be no mistake: I value the Open Access (OA )model of publication, for both moral and practical reasons that have been elaborated upon in many other places. Briefly: 1) OA is the right thing to do, as the results of publicly funded research should be available to the public and 2) it is the practical thing to, as the broadest sharing of knowledge possible is fundamental to educational and scientific advances.
This post deals with the difficulty that still exists on the ground. I see the current author-pays model of OA publishing as still somewhat problematic, with the result of driving many of my colleagues away from OA. One supporting argument for OA is that small universities and four-year colleges, and institutes in developing countries can ill-afford to subscribe to a large number of close-access publications. this places researchers and students at a disadvantage. Therefore, the OA model of publishing does them a favor by reducing subscription fees, granting broader access to publications.
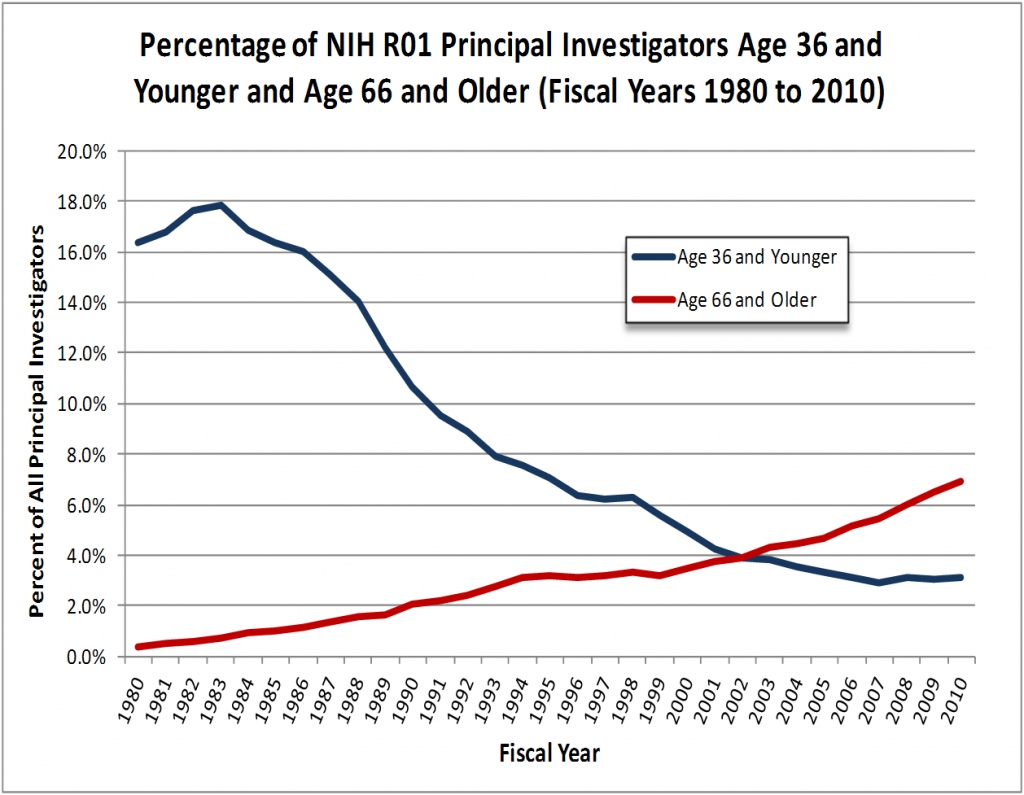
On the the flip side , it is in those very same institutes that researchers have less “disposable income” to pay for publications. $2500, a typical OA fee, for a lab funded by an R15 (small NIH grants given to less research-intensive institutes) or a small NSF grant is a larger chunk of change than $2500 for a lab holding a couple of R01s (the larger NIH “workhorse” grant). Knowing the limit on these grants, a researcher squeezed for funding would rather budget for an extra month for a graduate student than for OA publication fees. In way, OA fees are something of a regressive tax: it hurts those with less disposable income more. The OA advocates would say that the money saved by the institute from the reduction of library fees can be rolled into subsidizing publications. Some institutions do that by subscribing to OA journals, thus reducing the publication fees their authors are required to pay. However, many do not, and 10% of $2500 still leaves $2250 to pay.
Yes, PLoS grants “hardship” fee waivers, but many other publishers do not. However, requesting waivers in many cases is something of a dilemma: Prof. SmallU may have the $2500, but using those towards publication would mean running out of lab materials earlier than needed, or letting a graduate student off for the summer. In many cases it is not that the money is not there (after all, Prof. SmallU did manage to fund the research!) but facing this tough choice is problematic, and many people would be reluctant to ask for a subsidy or a waiver. Also, there is hardly any reward in small universities for publishing in OA. Publishing OA, by itself, figures very little, if at all in promotion & tenure decisions.
Therefore, when publishing those journals which have both options, OA and closed access, there is very little incentive to shell out the $2500 (usually more in the “two option” journals) once the paper is accepted. OA-only journals are often shunned altogether.
So what would be the solution? I agree with FakeElsevier that it has to come from the funding agencies. But maybe, instead of OA being a flat-fee, and hence regressive, it can be turned, with the help of the granting agencies, into a progressive fee. After all, those same agencies know how much funding a lab has anyhow, as this must be provided with every grant request, award, and progress report. If a lab is able to demonstrate a “publication hardship”, perhaps an extra subsidy can be given once a paper is accepted, provided it is used towards an OA publication. Knowing that this extra money is there may help nudge Prof. SmallU in the direction of publishing in OA. Also, the subsidy can be contingent upon the university subscribing to the OA journal, thus sharing the burden and creating and incentive for departmental library committees to pressure administrators to allocate funds towards OA access.
As it stands now, the motivation for low-budget labs, the supposed best beneficiaries of OA publishing, is not to publish OA. Unless stronger incentives are given, those labs will continue to get their reading material via those journals their library subscribes to, and through emailed electronic copies. Incidentally, a practice that the scientific publishing industry is starting to notice and is even attempting to stop.