In no particular order or ranking, recent and not-so-recent articles from PLoS-1. The common thread (if any): I thought they were pretty cool in one way or another.
1. Men don’t tell the truth about their penis. No kidding? But this is somewhat more serious. It has been accepted for some time that male circumcision dramatically reduces the rate of HIV infection. But recently, some reports have shown that high rates of infection prevail among circumcised men as well. But since circumcision is usually self-reported, could there be a problem there? This study shows that in a cross-sectional (sorry…) study among recruits to the Lesotho Defense Force, 50% of the men that reported they were circumcised were, in fact, partially (27%) or completely (23%) not circumcised. The researchers conclude that biases in the self-reporting of male circumcision may lead to erroneous reports that show high HIV infection rates among circumcised men.
Concluding quote:
…until further research can document improved methods for obtaining accurate self-reported MC [male circumcision I.F.] data, all assessments of MC and HIV prevalence, as well as projections for VMMC [voluntary male medical circumcision I.F.] interventions, should be informed by physical-exam-based data [as opposed toself reporting, I.F.].
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0027561
2. Share your data or GTFO.
Can sharing data help prevent errors and fraud?
From the abstract:
Background: The widespread reluctance to share published research data is often hypothesized to be due to the authors’ fear that reanalysis may expose errors in their work or may produce conclusions that contradict their own. However, these hypotheses have not previously been studied systematically
So Jelte Wicherts and his colleagues from the University of Amsterdam wanted to see whether sharing data was related to the number of statistical analysis errors in a paper. So, to phrase this as a null and alternative hypothesis:
H0:There is no difference in the number of statistical errors in those papers where the authors are willing to share data, and those where the authors are unwilling to do so.
H1: (one sided): the number of weaker evidence and statistical errors in papers where the authors are unwilling to share data is larger than those in which the authors are willing to share data.
Wicherts and colleagues contacted authors of 141 papers published in five journals of the American Psychological Association, requesting their data. Trouble is, they could not get enough authors to share data to make their own study significant: in a previous study, some 73% of the authors contacted were unwilling to share data. Wow.
However, authors publishing in two of these journals, Journal of Personality and Social Psychology (JPSP) and Journal of Experimental Psychology: Learning, Memory, and Cognition (JEP:LMC), were somewhat more forthcoming. Wicherts and colleagues therefore limited their analysis to a subset of 49 papers published in those journals. (Note that sometimes lack of data sharing is due to legitimate considerations, such as being part of an ongoing study, or third-party proprietary rights. However, those were not considerations in 49 papers analyzed here.)
Wicherts then checked for specific types of statistical errors in these papers, and compared the number of errors in papers from authors willing to share data to those who did not. Here are some of the findings:
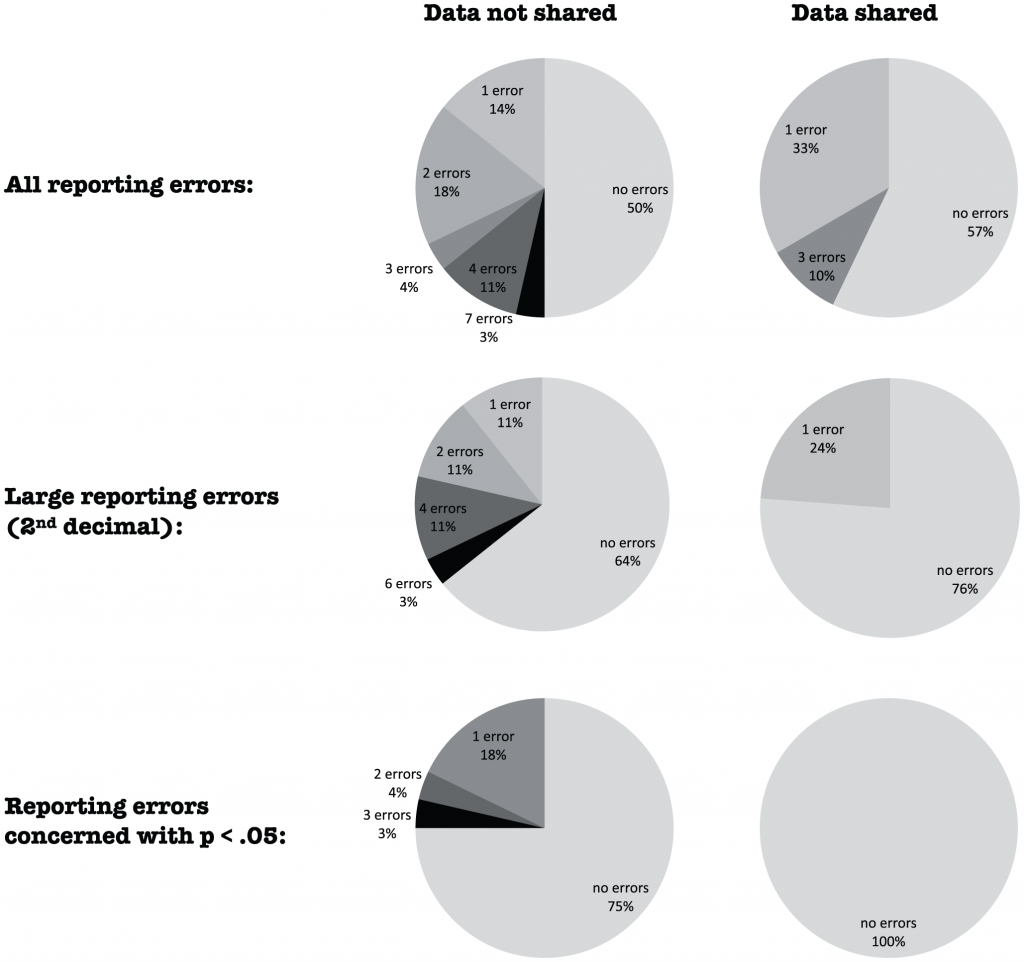
Distribution of the number of errors in the reporting of p-values for 28 papers from which the data were not shared (left column) and 21 from which the data were shared (right column) for all misreporting errors (upper row), larger misreporting errors at the 2nd decimal (middle row), and misreporting errors that concerned statistical significance (p<.05; bottom row). doi:10.1371/journal.pone.0026828.g001
Pretty clear picture: those papers where the authors authors were willing to share data were less prone to statistical errors.
Concluding quote:
In this sample of psychology papers, the authors’ reluctance to share data was associated with more errors in reporting of statistical results and with relatively weaker evidence (against the null hypothesis). The documented errors are arguably the tip of the iceberg of potential errors and biases in statistical analyses and the reporting of statistical results. It is rather disconcerting that roughly 50% of published papers in psychology contain reporting errors [33] and that the unwillingness to share data was most pronounced when the errors concerned statistical significance.
Although note that Wicherts is very careful about drawing conclusions:
Although our results are consistent with the notion that the reluctance to share data is generated by the author’s fear that reanalysis will expose errors and lead to opposing views on the results, our results are correlational in nature and so they are open to alternative interpretations. Although the two groups of papers are similar in terms of research fields and designs, it is possible that they differ in other regards. Notably, statistically rigorous researchers may archive their data better and may be more attentive towards statistical power than less statistically rigorous researchers. If so, more statistically rigorous researchers will more promptly share their data, conduct more powerful tests, and so report lower p-values. However, a check of the cell sizes in both categories of papers (see Text S2) did not suggest that statistical power was systematically higher in studies from which data were shared.
In fact, Wicherts also wrote a piece in Nature where he argued that sharing data can help avoid fraud, such as in the recent infamous case of Diederik Stapel, a highly regarded psychologist at Tilburg University in the Netherlands.
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0026828
3. Toilet paper. A study of surfaces of public restrooms has shown that they are covered with bacteria, mainly the kind that is known to live on and in humans. So now we have a somewhat broader view of the species living in restrooms, including the uncultured ones.
Two interesting quotes from the paper:
Although many of the source-tracking results evident from the restroom surfaces sampled here are somewhat obvious, this may not always be the case in other environments or locations.
Not sure about this bit: if the sources here are obvious, then is this paper a proof-of concept?
Also:
Unfortunately, previous studies have documented that college students (who are likely the most frequent users of the studied restrooms) are not always the most diligent of hand-washers.
No shit! (Pun intended).
Concluding quote:
Although the methods used here did not provide the degree of phylogenetic resolution to directly identify likely pathogens, the prevalence of gut and skin-associated bacteria throughout the restrooms we surveyed is concerning since enteropathogens or pathogens commonly found on skin (e.g. Staphylococcus aureus) could readily be transmitted between individuals by the touching of restroom surfaces.
Translation:
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0028132
Thomas, A., Tran, B., Cranston, M., Brown, M., Kumar, R., & Tlelai, M. (2011). Voluntary Medical Male Circumcision: A Cross-Sectional Study Comparing Circumcision Self-Report and Physical Examination Findings in Lesotho PLoS ONE, 6 (11) DOI: 10.1371/journal.pone.0027561
Wicherts, J., Bakker, M., & Molenaar, D. (2011). Willingness to Share Research Data Is Related to the Strength of the Evidence and the Quality of Reporting of Statistical Results PLoS ONE, 6 (11) DOI: 10.1371/journal.pone.0026828
Flores, G., Bates, S., Knights, D., Lauber, C., Stombaugh, J., Knight, R., & Fierer, N. (2011). Microbial Biogeography of Public Restroom Surfaces PLoS ONE, 6 (11) DOI: 10.1371/journal.pone.0028132