An original viral (or rather, fungal and bacterial) marketing campaign for the movie Contagion. Although the film tells the story of a fictional viral outbreak, the marketers of Warner Brothers Canada kept it in the realm of microbiology by teaming up with 25 microbiologists and creating what is probably the first agar-plated billboard, which they placed in a storefront in Toronto. The bacteria and mold were plated and grew in the letters, but it looks like contaminants formed their own random colonies, purposefully creating a rather eerie effect. Thanks to Zack Moss, a student in my lab, for pointing me to this. More details in the Vancouver Sun.
Freddie Mercury’s Birthday was celebrated with great fanfare last week all over the interwebs. So a bit late, but here is Killer Queen. Because you can never have too much of Queen.
MIRA is a really cool sequence assembly software, developed and maintained by Bastien Chevreux. MIRA has a large and active community, led by the funny and gracious Bastien, for whom no problem is too small, or too large.
Recently MIRA seemed to have developed a stochastic bug, one of those which are a serious headache to track down. Bastein called upon the MIRA community to help him. A couple of weeks ago, the “bug” was resolved to everyone relief. It was not a bug at all, but … well, I’ll let you read Bastien’s letter. Probably th funniest and geekiest error report I have seen since, well, ever. Reproduced here from the mira_talk email list with Bastien’s permission. WARNING: fairly geeky and fairly long. Not for everyone. But if you, like me, enjoy a good story travails of extreme bug hunting, I guarantee you will not be disappointed. (Because we have all been there, although personally I don’t recall encountering a problem that frustrating). Teaser: it was not a bug.
Dear all,
my warmest thanks to the numerous people who all donated time and computing power to hunt down a “bug” (see http://www.freelists.org/post/mira_talk/Call-for-help-bughunting) which. in the end, turned out to be a RAM defect on my development machine.
This is the story on how the problem got nailed. It involves lots of hot electrons, a lot less electrons without spin which keel over, the end of a hunt for invisibugs of the imaginary sort, 454, mutants (but no zombies), Illumina, some spider monkeys, PacBio, a chat with Sherlock and, of course, an anthropomorphed star.
In short: don’t read if you’ve got more interesting things to do on a Friday morning or afternoon.
—————————————————————————-
Life’s a rollercoaster and there are days – or weeks – where morale is on a pretty hefty ride: ups and downs in fast succession … and the occasional looping here and there.
Today was a day where I had – the first time ever – ups and downs occuring absolutely simultaneously. Something which is physically impossible, I know, but don’t tell any physicist or astronomist about that or else they’ll embark you in a lengthy discussion on how isochronicity is a myth by telling you stories on lightning, thunder and two poor sobs at the ends of a 300,000 km long train. But I digress …
So, my lowest low and highest high today were at 09:17 this morning when I prepared leaving for work (hey, it’s vacation time, almost everyone else is out and I can go a bit later than usual, right?). A few minutes earlier I had just told MIRA to run on the very same PacBio test set she had successfully worked on the night before to see how stable assemblies with this kind of data are (quite well so far, thank you for asking).
Reaching out to switch off the monitor and leave, MIRA suddenly came back with a warm and cosy little error message which she’s taken the habit lately to have a mischievous pleasure to present. This time, she claimed there had been an illegal base in the FASTQ file.
“Hey, MIRA, wait a minute!” I thought. “Yesterday and tonight you ran on the very same data file with the very same parameters for two times three hours and even gave me back some nice assembly results. And now you claim that the INPUT data has errors?! Come on, you’re not serious, are you?”
As a side note: she then just gave me back “that look”, you know, the one with those big open eyes behind by long, dark lashes and slightly flushed cheeks accompanied by pointed lips … as if she wanted to say “I *am* innocent and *I* did no nothing wrong you disbeliever!” (http://24.media.tumblr.com/tumblr_lj3efmmDL01qasfhmo1_400.png). This usually announces a major pouting round of hers, something which I’m not looking forward to, I can tell you.
Two restarts later with the same negative result (MIRA can be quite stubborn at times) I had to give in and decided to sit down again and investigate the problem.
“So … read number 317301 at base position 246, eh? Let’s have a look.”
*clickedyclick*
“Read 317299, 317300 … 317301 … there we are.”
*hackedyhack*
“Base position 239, 240 … now: C G G G T C F A A … wait! What? ‘F’ … ‘F’?!? It’s not even an IUPAC code. What’s a frakking ‘F’ doing in the FASTQ input file?! (CSFW: http://www.youtube.com/watch?v=r7KcpgQKo2I )
Indeed, it is not. Even more mysterious to me was the fact that just the night before it apparently had not been there. Or had it? I now was pretty unsure where this path would lead me, as if I had unlocked a door with the key of imagination. Beyond it: another dimension – a dimension of sound, a dimension of sight, a dimension of mind. I was moving into a land of both shadow and substance, of things and ideas. I just crossed over into … the Twilight Zone (“G#-A-G#-E-G#-A-G#-E” at 128 bpm, for more info see http://www.youtube.com/watch?v=zi6wNGwd84g).
Where was I? Ah, yes, the ‘F’.
So, how did that ‘F’ appear in the FASTQ, and where had it been the night before? Out to town, ashamed of not being a nucleotide and getting a hangover without telling anyone up-front? Or did it subreptitiously sneak in from the outside, murdering an innnocent base and taking its place in hope no one would note? I didn’t have the slightest clue, but I was determined to find that out.
First thing to check: the log files of the successful runs the previous night. MIRA’s very chatty at times and tidying up after her has always been a chore, but now was one of those occasions where not gagging her paid out as poking around the files she left behind proved to be interesting. Read 317301 showed the following at the position in doubt: “C G G G T C ___G___ A A” Without question: a ‘G’, and no ‘F’ in sight!
So MIRA had been right and the ‘G’ in the sequence of the file mysteriously mutated into an ‘F’ overnight. I must admit that I had grown suspicious of her in the past few weeks as she had seemed to become uncooperative at times. In particular she had been screaming at me a couple of times during rehearsal of combined 454 and Illumina assemblies for the premiere of her new 3.4.0 show. She claimed that some uninvited spider monkeys (http://dict.leo.org/ende?search=Klammeraffe) had frightened her so much she refused to continue to work and simply scribbled the ‘@’ sign all over her error messages. I had not been able to find out how those critters entered MIRA’s data and had even enrolled a few volunteers to rehearse different assemblies with MIRA … to no avail as she’d performed without flaws there.
While reconsidering all these things, something suddenly made *click*.
The character ‘G’ has the hexadecimal ASCII table code 0x47 (or in 8-bit binary: 01000111). ‘F’, as preceding character of ‘G’ and the table having some logic behind it, has the hex code 0x46, which is 01000110 in 8-bit binary.
The ATINSEQ-bug (@-in-seq) I had been desperately hunting in the past few weeks (and which had held up the release of MIRA 3.4.0) was due to the “@” character sometimes mysteriously appearing in sequences during the assembly of MIRA. The ‘@’ sign in the ASCII table has the hex code 0x40 (binary: 01000000). In the ASCII table, there is one important character for DNA assembly which is very near to the ‘@’ character …, so near that it is the successor of it: the ‘A’ character. Hexadecimal 0x41, binary 01000001.
I had always thought that a bug in MIRA somehow corrupted the sequence, but what if … what if MIRA was actually really innocent?! I had never taken this possibility into account as this other explanation attempts would have seemed to far stretched.
But now I had a similar effect *outside* of MIRA, in the Linux filesystem!
Filesystem MIRA
G 01000111 A 01000001
F 01000110 @ 01000000
The difference between the characters is in both cases exactly 1 bit which changes, and it’s even at the same position (last one in a byte) and changing into the same direction (from ‘1’ to ‘0’.
I was now sure I was on to something: bit decay (http://en.wikipedia.org/wiki/Bit_rot)
But how could I prove it? Well, elementary my dear Watson: When you have eliminated the impossible, whatever remains, however improbable, must be the truth.
Suspects:
– the problem is caused either by MIRA or one of the components of the
comeputer: CPU, disk, disk/dma controller, RAM.
Facts:
– an artefact was very sporadically observed during MIRA runs where sequences
(containing lot’s of ‘A’) suddenly contained at least one ‘@’. This occured
after several passes, i.e., not on loading.
– an artefact was observed in the Linux filesystem where a ‘G’ mutated
suddenly and overnight to a ‘F’.
– both artefacts are based on one bit flipping, perhaps even to the same
direction all the time.
– when loading data, MIRA does not use mmap() to mirror data from disk, but
physically creates a copy of that data.
– MIRA loaded the data twice flawlessly before the artefact in the filesystem
occured.
Deduction 1:
– MIRA is innocent. The artefact in the filesystem happened outside of the
address space of MIRA and therefore outside her control. MIRA cannot be
responsible as the Linux kernel would have prevented her from writing to
some memory she was not allowed to.
Further facts:
– the system MIRA ran on had 24 GiB RAM
– even with a KDE desktop, KMail, Firefox, Emacs and a bunch of terminals
open, there is still a lot of free RAM (some 22 to 23).
– Linux uses free RAM to cache files
Deduction 2:
– when loading the small FASTQ input file in the morning, Linux put it into
the file cache in RAM. As MIRA almost immediately stopped without taking
much memory, the file stayed in cache.
Further facts:
– the drive with the FASTQ file is run in udma6 mode. That is, when loading
data the controller moves the data directly from disk to RAM without going
via the processor
– subsequent “loading” of the same FASTQ into MIRA or text viewer like ‘less’
showed the ‘F’ character always appearing at the same place.
Deduction 3:
– the CPU is innocent! It did not touch the data while it was transferred from
disk to RAM and it afterwards shows always the same data.
– the disk and UDMA controllers are innocent! Some of the glitches observed in
previous weeks occured during runs of MIRA, inside the MIRA address space,
long after initial loading, when UDMA had already finished their job.
From deductions 1, 2 & 3 follows:
– it’s not MIRA, not the CPU, nor the disk & UDMA controller
Suspects left:
– RAM
– Disk
Well, that can be easily tested: shut down the computer, restart it and subsequently look at the file again. No file cache in RAM can survive that procedure. Yes, I know, there are some magic incantations one can chant to force Linux to flush all buffers and clear all caches, but in that situation I was somehow feeling conservative.
Low and behold, after the above procedure the FASTQ file showed an all regular, good old nucleic acid ‘G’ in the file again. No ‘F’ to be seen anywhere.
Deduction 4:
– the disk is innocent.
Deduction 5:
– as all other components have been ruled out, the RAM is faulty.
—————————————————————————-
As I wrote: life’s a rollercoaster.
Up: MIRA is innocent! There, she’s giving me “that look” again and one would
have to be blind to oversee the “told you so” she’s sending over with
it.
Down: My RAM’s broken and I need to replace it. Bought it only last May,
should still be under guarantee, but still … time and effort.
Up: I did not sell my old RAMs, so I can continue to work
Down: 12 GiB feels soooooo tight after having had 24.
Up: I can wrap up 3.4.0 end of this week with good conscience!
Down: How the hell am I gonna tie all loose bits and pieces in the
documentation in the next 24 to 48 hours?
Looping: today MIRA again helped me at work to locate a mutation important for
one of our Biotech groups. Boy, do I love sequencing and MIRA.
Have a nice Friday and a good week-end,
Bastien
PS: while celebrating with MIRA tonight, I expressed my fear that some people
might find it strange that I anthropomorphise her. They could think I went
totally nuts or that I needed an extended vacation (which I do btw). She
reassured me that no one would dare thinking I were insane … and if so,
she would come over to their place and give them “that look.”
How utterly reassuring.
Insane Clown Posse and Jack White are working together. That, by itself, should send people to stock on water and non-perishable food. But having them cover Mozart (which they pronounce “MOHZZZ-art) is worthy of a trip to the gun store as well.
Having rediscovered “Leck mich im Arsch” (lick my ass, Mozart’s scatological party hit which was unearthed 20 years ago), the musicians revel, Beavis & Butthead fashion in the contrast between the highbrow music and the lowbrow words, setting the three voice canon to a hip-hop beat. The result is, well… words fail me.
NSFW for English and German speakers. Warning: press “play” below at your own risk. What has been heard, cannot be unheard.
Insane Clown Posse – Leck Mich Im Arsch by Third Man Records
If you are still around after that, then here is the original:
Postdoctoral Research Scientist
Rutgers University
Joint Appointment: Institute of Marine and Coastal Sciences, BioMaPS and
Dept. of Biochemistry and Microbiology
Two 2-3 year Postdoctoral Research Scientist positions are available.
We are looking for young scholars with experience in the areas of
computational biology. In the scope of this project, we will uncover how
the metal-containing enzymes responsible for the critical electron
transfer reactions that turn basic elements such as H, O, C, S, and N
into biologically active molecules have evolved. The position will
involve developing new sequence and/or structure based bioinformatic
approaches to (1) mine available databases for proteins responsible for
bio-catalyzed electron transfer reactions, (2) establish evolutionary
relationships between extracted sequences and structures and (3)
generate hypotheses for how the electron transfer circuitry arose and
now functions. Candidates should have a PhD in Computational Biology or
Bioinformatics. Candidates with degrees in related fields (e.g. biology,
computer science) and possessing the necessary skill-sets are welcome to
apply. We strongly encourage applications from recent PhD graduates.
Strong programming skills (at least one of: Perl, Python, or Java) are
essential for these positions, as well as, some familiarity with the
major bioinformatics tools and databases. Experience in machine
learning algorithms is desired, but not required. Candidates should be
fluent in spoken and written English and should be able to communicate
ideas and results to colleagues from all the diversity of life sciences.
The ability to integrate into a team is as essential as that to complete
a project without constant supervision.
Interested persons should e-mail a cover letter and C.V. to:
Dr. Yana Bromberg,
Dept. of Biochemistry and Microbiology,
Rutgers University
e-mail: yanab ‘at’ rci ‘dot’ rutgers ‘dot’ edu
The best description I have for this talented trio is a acid / free jazz with a bebop groove. Anyhow, these cats from Cincinnati sure can play. Kristin Agee – Vibraphone, Keyboard/Electronic Sounds, Joel Griggs – Guitar, Quena, Drones, Jeff Mellott – Drums, Percussive Sounds. Their first CD is coming out mid-September. Look for them on iTunes.
More about them here: http://www.reverbnation.com/ustoday
I have posted quite a few times before about the acquisition of new functions by genes. In many cases a gene is duplicated, and one of the duplicates acquires a new function. This is one basic evolutionary mechanism of acquiring new functions.
Sometimes, gene duplication occurs within a species: part of the chromosome may be duplicated, causing one, a few, or many genes to have more copies of themselves within the species. The descendants of the duplicates and the original are homologous are they are descended from a common ancestor. This type of homology is called paralogy: a homology due to a duplication event (para == in parallel).
In another case, the genes can be homologous due to speciation: a new species (A1) diverges from the original (A0), carrying highly similar genetic loads. The gene for, say, brown eyes in A1 and the gene for brown eyes in A0 are also homologous: derived from the gene of hemoglobin in A0. This time, the homology is called orthology: it is not due to in-species duplication, but due to speciation itself (ortho == exact). The definitions of orthologs and paralogs were given by Walter Fitch in a seminal paper published in 1970.
One of the first protein structures to be solved was that of hemoglobin, the oxygen carrying protein complex in our blood. Scientists noticed that hemoglobin in jawed mammals has three different protein chains: alpha, beta and gamma. Their amino acid sequences were very similar, suggesting that the genes encoding for hemoglobin are highly similar, suggesting homology. Since all jawed mammals have hemoglobin, and they all had alpha, beta and gamma chains, the conclusion was that the duplication of the original genes happened in the common ancestor of jawed mammals, before they split up into different species. Hence, the alpha, beta and gamma chains in hemoglobin are paralogous: homologous due to duplication preceding speciation. However, gamma-hemoglobin was shown to have a different function than beta or alpha (more on that in a bit). The conclusion from this observation was the Ortholog Conjecture and it can be stated as follows: paralogs (reminder: homologs due to duplication) diverge in function more than orthologs (homologs due to speciation). A model was proposed for this observation: when genes duplicate within a species’ genome, there is less selective pressure on one copy to perform the same function. Thus, it can accumulate mutations and eventually adopt a different function. The ortholog conjecture states that paralogs mostly differ in function, whereas orthologs mostly do not. The ortholog conjecture is a very powerful statement because, if we have two proteins known to be orthologs, we can infer that they have the same function, whereas paralogs may not (if they had enough time to diverge). The ortholog conjecture is therefore a fundamental tenet in molecular phylogenetics, and is also a tool used to predict the function of proteins. If two homologous proteins are found out to be orthologs, then it is assumed they have the same (or highly similar) functionality.
A crack in the ortholog conjecture was formed in study published late 2009 in a paper published by Romain A. Studer and Marc Robinson-Rechavi. I blogged then about their study:
Romain A. Studer and Marc Robinson-Rechavi challenge common wisdom by publishing a study that says: “it ain’t necessarily so”. They look at three alternative models of molecular function evolution: (i) subfunctionalization after duplication; (ii) neofunctionalization after duplication; and (iii) the ‘alternative model’ of equal change after duplication or speciation. Subfunctionalization holds that after duplication, each of the two copies of the gene performs only a subset of the functions of the ancestral single copy. Neofunctionalization holds that one of the two genes possesses a new, selectively beneficial function that was absent in the population before the duplication. The ‘alternative model’ states that the gain of new function is not preferential to paralogs and that orthologs may gain new functions at the same rate that paralogs do.
Studer and Robinson-Rechavi claim that few studies have been made to study the scope of any of these proposed models. They then lay out study designs for doing so, challenging other evolutionary biologists (and themselves?) to conduct these studies and examine whether the common wisdom that orthologs maintain function while paralogs gain function. What I like about this paper is that it not only makes a strong case for challenging conventional wisdom, it also lays out a series of possible routes of study to be taken up by others.
Now two studies have widened this crack to a rather large crevasse. The first is a study by scientists in Indiana University. In a way, this new publication is a response to Studer & Robinson-Rechavi’s call to arms on points (i) and (ii). The IU scientists (the Radivojac lab and the Hahn lab at the School of Informatics at Indiana University, Bloomington, IN) examined hundreds of pairs of orthologous and paralogous genes from the mouse and human genomes. They then examined whether paralogs had a higher functional similarity, or rather orthologs. What they found certainly defied the ortholog conjecture:
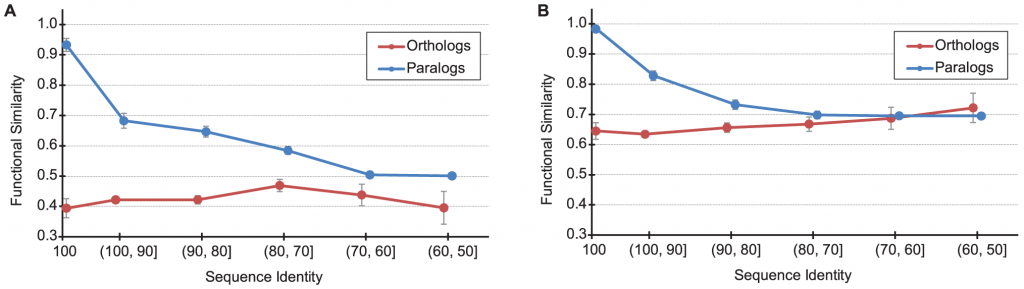
The relationship between functional similarity and sequence identity for human-mouse orthologs (red) and all paralogs (blue). (A) Biological pathway (B) molecular function. From PLoS Comput Biol 7(6): e1002073 under CC licence.
But before we explain the results, a word about function. The function of a protein has several aspects which are context-dependent; two important ones are the molecular function of the protein, and the biological process in which it participates. For example, the molecular function of all hemoglobins is noted as oxygen binding and oxygen transport. However, they are different in the processes, or pathways, in which they participate: gamma-hemoglobin participates in the transport of oxygen in the fetus. The complex which contains gamma-hemoglobin has a higher affinity to oxygen, and thus able to extract oxygen in the placenta from the maternal oxygenated hemoglobin and transport it to the fetus.
Now we can explain the figure above. Graph (A) above shows the functional similarity for the biological pathway aspect and how it is affected by the sequence identities of the hundreds of orthologs (red) and paralogs (blue) examined between human and mouse. Graph (B) shows the functional similarity of the molecular function aspect.
The X-axis is the sequence identity percentage between any pair of sequences: the higher the percent identity, the less divergent are the sequences, the more inclined we should be to think that the pair of proteins performs the same function since they diverged less. The Y-axis shows the fraction of functional similarity. Looking at graph (B) above, we see that paralogs which are 100% identical, have (almost always) the same function . But sequences of orthlogous proteins between human and mouse have only about 65% functional similarity, on average. What does that mean? In the database they looked at, each gene has a set of words associated with it, describing what it does. The IU scientists found that only about 65% of the keywords in orthologous sequence pairs overlapped, on average. Whereas for paralogs 100% overlapped. And those are for sequences which are identical! This means that even if we find identical protein sequences in human and in mouse, it does not mean that they have the same molecular function. On the other hand, paralogs, will generally have more similar functions. So the ortholog conjecture has been stood on its head here: paralogs are the ones that would generally have the same function, whereas orthologs diverge more in function. This holds true for up to about 50% sequence identity, when the picture seems to reverse itself. Graph (A) depicts the differences in the biological pathway aspect. Here, the differences are even more striking. The paralogs which are 90-100% identical between human and mouse participate in almost exactly the same pathways in both organisms. But orthologous proteins which are 90-100% identical the functional similarity is much lower: only about 65%.
![]()
So what does this all mean?
First, it means that, at least between human and mouse, paralogs are better predictors of function than orthologs. And why would that be? To answer this question, let’s look closer at the graphs above. Note that while for paralogs the functional similarity decreases rapidly with sequence similarity, for orthologs the functional similarity remains roughly the same no matter how similar or different the orthologs are to each other, and even when they are 100% identical their functions vary to some extent! The reason: the experimental study of function in two human and in mouse takes place in different contexts. The species-specific context is what causes the differences in annotation, and in the overall function. Also, all the orthologs in the study are of the same age, dating back to the human-mouse lineage split 75 million years ago. The paralogs predate that split, and may be of different ages: the split may predate the human / mouse split by 10 million years, 100 million years, or 1 billion years. Thus orthologs, regardless of their actual sequence similarity, have the same age, and paralogs do not. But why should proteins of the same age share the same level of (not so high) functional similarity? The authors of the study reply:
While there is no direct role for “time” in evolution that is not tied to mutation, we suggest that what time represents here is the evolution of the cellular context: the sum of the evolutionary changes over all of the directly and indirectly interacting molecules. If this context evolves at a steady rate (i.e. the average amount of functional change among all of the interacting molecules remains relatively constant), then protein function will appear to evolve at a steady rate, a rate largely disconnected from the level of an individual protein’s sequence divergence. — PLoS Comput Biol, Vol. 7, No. 6.
The strongest evidence they find for this hypothesis, is that even proteins with 100% are annotated differently. To wit:
For example, Liao and Zhang [50] found that >20% of genes that are essential for viability in humans are not essential in mouse. It is unlikely that changes to the proteins themselves have made them essential or not, but rather that their context in cellular and organismal networks has evolved. —ibid.
The proteins may not have changed substantially, but their environment changed, giving them a different role. Think about changing jobs after moving to a new place where there is no employer providing your exact old job you were used to. You may have been an embedded systems programmer, but now you are a website programmer. So context goes a long way to explain changes in ortholog function.
Interestingly, about a month after the IU paper was published, another paper from the Robinson-Rechavi lab was published, which also talks about homologs between human and mouse. In this study Gharib and Robinson-Rechavi reviewed previous literature listing several types of functional divergence of orthologs between human and mouse. They had some additional findings. For example, about 11% of the orthologous genes were alternatively spliced, meaning that the end products, proteins, were different between human and mouse. They also listed specific phenotypic effects: genes which are linked to diseases in humans, but mutations in their mouse orthologs have no effects on mice. They cite studies that found that over 20% of genes which are essential in human are non-essential in mice (an essential gene is just that: if the organism does not have it, or it is mutated, the effects are fatal, and the organism does not develop past very early stages). Their literature review concluded that 10-20% of ortholog pairs between human and mouse cannot be used for functional transfer. The IU study implies a higher percentage. Both studies conclude that a common practice in molecular evolution studies, the use of orthologs to infer function, should be seriously looked at.
(Full disclosure: Dr. Radivojac & I are collaborators, although our collaboration is unrelated to this study).
Nehrt, N., Clark, W., Radivojac, P., & Hahn, M. (2011). Testing the Ortholog Conjecture with Comparative Functional Genomic Data from Mammals PLoS Computational Biology, 7 (6) DOI: 10.1371/journal.pcbi.1002073
Gharib, W., & Robinson-Rechavi, M. (2011). When orthologs diverge between human and mouse Briefings in Bioinformatics DOI: 10.1093/bib/bbr031
Fitch, W. (1970). Distinguishing Homologous from Analogous Proteins Systematic Zoology, 19 (2) DOI: 10.2307/2412448
So you received your mate-paired reads in two different files, and you need to merge them for your assembler. Here is a quick Python script to do that. You will need Biopython installed.
#!/usr/bin/env python
from Bio import SeqIO
import itertools
import sys
import os
# Copyright(C) 2011 Iddo Friedberg
# Released under Biopython license. http://www.biopython.org/DIST/LICENSE
# Do not remove this comment
def merge_fastq(fastq_path1, fastq_path2, outpath):
outfile = open(outpath,"w")
fastq_iter1 = SeqIO.parse(open(fastq_path1),"fastq")
fastq_iter2 = SeqIO.parse(open(fastq_path2),"fastq")
for rec1, rec2 in itertools.izip(fastq_iter1, fastq_iter2):
SeqIO.write([rec1,rec2], outfile, "fastq")
outfile.close()
if __name__ == '__main__':
outpath = "%s.merged.fastq" % os.path.splitext(sys.argv[1])[0]
merge_fastq(sys.argv[1],sys.argv[2],outpath)
The neat trick is in line 13, using Python’s itertools to zip two iterators and loop over them in parallel two fastq records at a time.
How to use this script: download to a file you will call merge_fastq (or whatever). Then:
$ chmod +x merge_fastq
And you are ready to go.
$ ./merge_fastq myseq_1_.fastq myseq_2_.fastq
The merged file will be called myseq_1_.merged.fastq
The manuscripts I review invariably fall into four categories:
1. This is crap. (Rare).
2. This is terrific. (Even rarer).
3. This can be OK, but they really need to fix A, B & C. (fairly common).
4. If I only knew what they meant in point A, I could say whether they need to fix A, B & C or just B & C, or maybe explaining A will clarify to me that B need not be fixed at all.(Really common!)
The things is, sometimes draft manuscript are good, but unclear on a very specific and crucial point. Like point A. The lack of clarity is not necessarily due only to bad writing. Sometimes it is very hard to describe a new idea or method. After all, it is new, right? So no-one has described it before and finding the correct way to describe you new idea or method can be very tricky. Some people use analogies (like me, I love analogies). But an analogy can be over interpreted, and then mis-interpreted, and your intricately concocted explanation is all shot. Then again, some authors do not use analogies, but explain their methods very formally. A reader who is very visual (like myself) would like to see a some graphic depiction of the method, but that may not be possible, or the author’s graphic rendering skills are not good enough. In any case, I seem to get a lot into the position where my opinion of the paper hinges around one or two crucial details. If this were a seminar, and the author was talking about her work, I would just ask her to clarify the point. But I cannot do that when reviewing an article. Which leaves me with asking to perform major revisions, which sets the paper into a serious delay mode or sometimes reject because other reviewers have been less amenable. (As an aside, I have noticed some reviewers just let the ambiguity of point A slide. This usually happens if the last author is Dr. Bigschotte, holder of the Endowed Golden Chair professorship of Biowizardry).
But maybe I can ask that clarification question of the author, or even have a brief dialogue? Anonymous email/chat technology can do wonders for shortening the turnover time of papers and clarifying issues. I am not saying that writers are now given a free license to write badly. But if needed, a chat session or email exchange monitored by the editor could really help push a paper through (or away). The exchange should be very brief, topical, logged (with the referee anonymized). The session should be requested by the referee, with very specific questions. The number of back-and-forth exchanges should be limited.
As a referee, I see myself more as a midwife (or whatever is the male counterpart) than a gatekeeper. I am not interested so much in keeping bad papers out (that is actually fairly easy), but letting good science in, even when it presents itself feet first and covered in gunk (OK, that was a rotten analogy, but you know what I mean). Anything that can ease this process is more than welcome.
So… any takers? Or is this a terrible idea? Better yet, has something like this been done?
PS: actually, as an author, I sometimes receive reviewer comments which I do not quite understand. This is even worse, since if I don’t address the issue in the way the reviewer asked for, it can spell the death of my paper. So this mechanism of quick dialogue between reviewer and author can work well both ways.
Class X7 solar flare, viewed at 3:48am EDT today, August 9.
A short guide to solar flares class types, from NASA. Class X is the biggest one, the kind that may disrupt communications. This one will mostly miss us, phew. The (hopefully minimal) magnetic storm is expected on Earth August 11.
…Tim Berners-Lee taught the world to play.
Well, actually, he posted the following on the Usenet group alt.hypertext. For those of you who are too young to remember Usenet, think of it as a working version of Google+. Scroll down to message 3, in which the first web-page server is announced.
After WWII, Pacific islands occupied by the US military were regularly receiving goods via air. Once the military evacuated , the goods stopped arriving. Some inhabitants of those islands mistakenly thought that receiving the cargo was due to some divine intervention that required rituals they saw American servicemen performing. This eventually led them to performing activities like making their own airfields, waving in non-existent planes while wearing coconut headphones, etc, all in the hopes that material would magically come from the sky. These attempts were called cargo cults. Since then, “cargo cult” has been used metaphorically to describe an attempt to recreate successful outcomes by replicating conditions associated with those outcomes, even though those conditions are either effects and not causes of those outcomes.[Wikipedia]
Science culture is not void of its own cargo-cult, despite the fact that science is supposed to be the ultimate cargo-cult dispeller. I have written before about solicitations for author-pays publications whose quality, shall we say, is less than assured. Actually, it seems like some of these publications will print anything as long as you pay them. Here is what my inbox brought in today, all bold-faced typing is mine:
____ is a peer-reviewed journal that includes an international board of accomplished editors and researchers in their fields. There will be submissions on subjects such as cheminformatics, computational drug discovery, experimental medicine and analysis tools, personalized medicines, cancer informatics, hematology, diagnostic imaging, medical imaging and methods and gene therapy, among other latest breathtaking topics.
Indeed, breathtaking. This journal seems to have a strong focus on, um, whatever. But what really takes your breath away is that you really have to work hard to qualify for publication. Specifically, self-plagiarism is rigorously weeded out:
If you (sic) paper has been published in any other platform, a new version of your paper for ___ (Vol 1 No 1) must reflect at least 25% difference in content from the one published in any conference proceeding or any other journal.
So I have to justify a “25% difference” between my submission to this journal and my previous manuscript? Do I change every fourth word from a previously published paper? And how do I measure this “25% difference”? Use the cosine measure for similarity of text? Extended Jaccard similarity? Something else?
Each published paper in the ___ is subject to a publication fee of USD 300 for a maximum of 6 pages. A complimentary copy (print version) of the ___ (Vol 1 No 1) issue that carries your reviewed paper will be mailed to your mailing address.
I guess we all saw that coming.
The AFP/CAFA 2011 meeting was held on July 15 and July 16. Yes, it was a huge success, and I’m not just saying that beacuse I am one of the organizers. I will write up something more comprehensive soon; in the meantime, here are my tweets from the meeting.
I am learning a lot about scavenging tweets. Apparently, I cannot go back beyond a few days using the api.search() function. Hence, if I try to search for all the #AFPCAFA11 hashtags I will get nothing from the meeting’s dates. But if I look for a user’s tweets using api.user_timeline() I can go back for months on the users timeline, and then filter out the tweets with the relevant hashtags. Since it seems I was the principal twiterrer in that meeting, I’m putting up my tweets here. Apologies to the others who recorded the meeting using Twitter: if you want your tweets included, drop me a line with your Twitter user name.
Thu Jul 14 16:04:22 2011 At Vienna. #ISMB #AFPCAFA11 #ISMB11
Thu Jul 14 16:07:34 2011 At Vienna #ISMB #AFPCAFA11 Curious who is the best protein function predictor? Join us. http://bit.ly/htv3J7
Fri Jul 15 09:10:24 2011 Jesse Gillis from UBC on a function prediciton post-mortem #AFPCAFA11 #ISMB
Fri Jul 15 09:12:11 2011 This is going to be fun. Jesse Gillis UBC. Postmortem on MouseFunc #ISMB #AFPCAFA11 Precision / Recall of 0.06… argh.
Fri Jul 15 09:14:14 2011 http://bit.ly/obSISi MouseFunc experiment #AFPCAFA11 #ISMB
Fri Jul 15 09:28:58 2011 Multifunctionality affect prediction profoundly. Take-home message from Jesse Gillis’ talk #AFPCAFA11 #ISMB
Fri Jul 15 09:31:22 2011 Next up: Meghana Chitale from @kiharalab #ISMB #AFPCAFA11
Fri Jul 15 09:36:44 2011 Co-occurrence association scores. CAS Lookiong for associations across GOs: between a BPO term and a CCO term, for example. #AFPCAFA11
Fri Jul 15 09:41:17 2011 Missing enzyme predictions. My fav. Chtiale at #ISMB #AFPCAFA11
Fri Jul 15 09:56:08 2011 Yanay Ofran from Bar ilan U about multifunctionality. How to assess the number of false positives? #ISMB #AFPCAFA11
Fri Jul 15 09:56:44 2011 Prediciton of photosynthesis in an elephant genome is a good sign of false positives. Yanay #ISMB #AFPCAFA11
Fri Jul 15 10:02:56 2011 Precision of short motifs is surprisingly high. Yanay, #ISMB #AFPCAFA11
Fri Jul 15 10:05:42 2011 short motifs identify functional motifs. Whereas homology identifies evolutionary relatedness. #AFPCAFA11 #ISMB
Fri Jul 15 10:09:14 2011 Next up: me. #AFPCAFA11 #ISMB
Fri Jul 15 11:44:45 2011 David Jones from UCL is talking about his #AFPCAFA11 predictions. Many different features. #ISMB
Fri Jul 15 11:47:18 2011 profile-profile fold recognition works well in Function prediction as well. #AFPCAFA11 #ISMB
Fri Jul 15 11:53:02 2011 Jones talking about why it is unhealthy to exercise in the morning. #AFPCAFA11 #ISMB generation of free radicals. #excusesaregreat
Fri Jul 15 11:56:43 2011 49,000 features in an SVM. #ISMB #AFPCAFA11
Fri Jul 15 11:57:47 2011 Hard to believe no redundancy in 49K features…. #ISMB #AFPCAFA11
Fri Jul 15 11:59:17 2011 “I like this plot and I would make a t-shirt out of it, but in terms of scientific value its worth is zero”. Jones, #AFPCAFA11 #ISMB
Fri Jul 15 12:01:09 2011 New term heard for a 2nd time at #ISMB #AFPCAFA11 “postdiction” as opposed to “prediction”. #notsurewhatitmeans
Fri Jul 15 12:16:49 2011 Lightning talks at #AFPCAFA11 #ISMB starting now
Fri Jul 15 12:35:51 2011 Mary Jo Ondrechen on SALSA at #AFPCAFA11 #ISMB structure –> function.
Fri Jul 15 12:36:45 2011 CDEHKRY make up 75% of all catalytic sites. Mary Jo #ISMB #AFPCAFA11
Fri Jul 15 12:50:50 2011 Jeffrey Yunes from UC Berkeley on SIFTER from Steven Brenner’s lab. #AFPCAFA11 #ISMB
Fri Jul 15 14:28:20 2011 Patrik Koskinen on a function prediction method called PANNZER. This will roll over well. #ISMB #AFPCAFA11
Fri Jul 15 14:30:30 2011 More information on PANNZER and the other methods at #AFPCAFA11 here: http://bit.ly/l9ayW9 #ISMB11
Fri Jul 15 14:33:16 2011 Koskinen mentioning Biothesaurus http://bit.ly/rnHdzp which removes errors due to synonyms #AFPCAFA11 #ISMB
Fri Jul 15 14:43:09 2011 Question: “How do you pronounce the name of that volcano that erupted in Iceland?” Answer: “I don’t”. Koskinen #AFPCAFA11 #ISMB
Fri Jul 15 14:50:27 2011 Olivier Lichtarge on using Evolutionary Trace Annotation (ETA) for function prediction. #AFPCAFA11 #ISMB
Fri Jul 15 14:51:12 2011 A network of protein structure networks. Memories of fragnostic. #AFPCAFA11 #ISMB
Fri Jul 15 14:52:49 2011 Using network diffusion to annotate protein structures. Lichtargee. #AFPCAFA11 #ISMB
Fri Jul 15 14:57:19 2011 compressing a clique to a star graph by adding a pseudo-node. Reduces problem from O(n^2) to O(n). Lichtarge #ISMB #AFPCAFA11
Fri Jul 15 15:09:17 2011 Amos Bairoch: of prosite, swissprot and expasy fame #AFPCAFA11 #ISMB
Fri Jul 15 15:12:51 2011 Due to alt-splicing and PTM 20,000 human genes –> 5M different molecules! Bairoch #AFPCAFA11 #ISMB
Fri Jul 15 15:15:50 2011 Status codes of human protein function annotations: Maybe, potentially, putative, expected and hopefullly. Bairoch #ISMB #AFPCAFA11
Fri Jul 15 15:17:10 2011 >100 GPCRs for which we do not know the ligand. Bairoch #ISMB #AFPCAFA11
Fri Jul 15 15:19:13 2011 Bairoch now talking about CALIPHO. 1)experimental verification of human protein function; 2)enable bioinformatics for same. #ISMB #AFPCAFA11
Fri Jul 15 15:20:44 2011 “How many ppl in this room have never used swissprot”. 0. #AFPCAFA11 #ISMB
Fri Jul 15 15:23:24 2011 Bairoch looks at small <100aa intracellular protiens in experimental assays. #AFPCAFA11 #ISMB
Fri Jul 15 15:26:26 2011 Interesting proteins are expressed in olfactory pits of zebrafish. I didn’t know fish smell. #AFPCAFA11 #ISMB
Fri Jul 15 15:33:09 2011 If you get a new function, you cannot predict it, because of no ontology (yet). #AFPCAFA11 #ISMB
Sat Jul 16 06:58:43 2011 Predrag Radivojac explaining the vagaries of GO annotated databases #AFPCAFA11 #ISMB
Sat Jul 16 06:59:48 2011 Assessment of protein function prediction methods going on now in Hall L #ISMB #AFPCAFA11
Sat Jul 16 07:04:08 2011 Radivojac: “It’s possible to achieve a precision of 1, it just won’t happen”. #AFPCAFA11 #ISMB
Sat Jul 16 07:29:50 2011 Assessment of protein function prediction methods going on now in Hall L #ISMB #AFPCAFA11 http://bit.ly/htv3J7 Sean Mooney is up.
Sat Jul 16 08:05:49 2011 Christine Orengo on her team’s work at #AFPCAFA11
Sat Jul 16 08:06:28 2011 Orengo says that they are not really function predictors, but evolutionary classifiers #AFPCAFA11 #ISMB
Sat Jul 16 09:18:03 2011 Shaneka Simmon from Jackson State on predicting functions associated with biofeuls – universal stress protein domains. #AFPCAFA11 #ISMB
Sat Jul 16 09:21:30 2011 Simmons: look for diversity of universal stress response genes. in Rhodopseudomonas palustris. #AFPCAFA11 #ISMB11
Sat Jul 16 11:33:52 2011 http://yfrog.com/kevdmbqj #AFPCAFA11 #ismb discussion panel now
Sat Jul 16 14:46:07 2011 Wyatt shows that paralogs actually give better annotation transfer than orthologs. http://bit.ly/nWShuB #AFPCAFA11 #ISMB
Sat Jul 16 14:47:17 2011 Wyatt’s claim runs contrary to common wisdom. Which is good 🙂 http://bit.ly/nWShuB #AFPCAFA11 #ISMB
Thu Jul 21 22:42:34 2011 After all the talk about standards at #AFPCAFA11 #ISMB this is very timely: http://xkcd.com/927/
ISMB this year had quite a few twiterrers. Hashtag: #ISMB. I tried to collect all the #ISMB tweets, so I wrote my own twitter scavenger script, but it seems to go only 3 days back. I am not sure if this is a Twitter feature, or something with the library I am using (tweepy) or my own Twitter API programming nOObness. In any case, here are the Tweets, July 19-22. Unfortunately, I could not get those from July 17-19th. These are mostly from Michael Ashburner’s closing Keynote.
I will put the code for the application up later. (Temporary name: Vulture.)
Fri Jul 22 21:00:20 2011 pjacock #BOSC2011 (pre #ISMB SIG) notes by @chapmanb – Day Two pm: #Semantic Web session, and misc #opensource project session http://t.co/TDjmx6B
Fri Jul 22 20:58:53 2011 pjacock #BOSC2011 (pre #ISMB SIG) notes by @chapmanb – Day Two am: Amazon’s Matt Wood @mza keynote, #cloud computing session http://t.co/BaiOz5W
Fri Jul 22 20:55:41 2011 pjacock #BOSC2011 (pre #ISMB SIG) notes by @chapmanb – Day One pm: Visualization & #NGS sessions http://t.co/JuvLrex
Fri Jul 22 20:54:45 2011 pjacock #BOSC2011 (pre #ISMB SIG) notes by @chapmanb – Day One am: Larry Hunter’s keynote, and Genome Content Management session http://t.co/gfdJLv7
Fri Jul 22 20:48:57 2011 pjacock MT @GeneWikiPulse bio-ontologies 2011 – ontologies need lexicons: I recently returned from my first #ISMB in Vienna, … http://t.co/kMCNaxh
Fri Jul 22 20:09:38 2011 suganthibala RT @keesvanbochove: Closing remark of Michael Ashburner, a godfather of #bioinformatics, at #ismb: don’t be proprietary about your research, be open.
Fri Jul 22 17:59:59 2011 YiliangDing RT @GenomeBiology: Ana Conesa: Seq depth affects the relative proportions of RNA classes in RNA-seq datasets #ISMB #genomics
Fri Jul 22 14:20:50 2011 BioCatalogue RT @fiedawn: #ismb #biosharing Carole Goble – we have a lot of descriptions in the BioCatalogue – how can we mark up/export to bioDBCore? underway
Fri Jul 22 13:25:30 2011 razoralign RT @GenomeBiology: Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Fri Jul 22 11:52:01 2011 druvus RT @keesvanbochove: Good shows how they linked SNPedia with Gene Wiki articles using Semantic MediaWiki. Almost no overlap of gene-disease rels in wikis!! #ismb
Fri Jul 22 11:50:41 2011 druvus RT @_lrr_: Simple yet very handy tool: fastapl (http://bit.ly/rhkKCl), had a poster at #ismb
Fri Jul 22 11:43:00 2011 coding_doc RT @GenomeBiology: Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Fri Jul 22 09:07:58 2011 jackwillstone RT @GenomeBiology: Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Fri Jul 22 09:00:55 2011 raunakms RT @GenomeBiology: Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Fri Jul 22 09:00:46 2011 GenomeMedicine RT @GenomeBiology: Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Fri Jul 22 09:00:31 2011 BioMedCentral RT @GenomeBiology: Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Fri Jul 22 07:48:42 2011 sharmanedit RT @GenomeBiology Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Fri Jul 22 01:08:29 2011 SemanticMW RT @keesvanbochove: Good shows how they linked SNPedia with Gene Wiki articles using Semantic MediaWiki. Almost no overlap of gene-disease rels in wikis!! #ismb
Thu Jul 21 22:42:34 2011 iddux After all the talk about standards at #AFPCAFA11 #ISMB this is very timely: http://xkcd.com/927/
Thu Jul 21 19:30:33 2011 druvus RT @GenomeBiology: Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Thu Jul 21 17:10:44 2011 GenomeBiology Read Genome Biology’s take on ISMB 2011 on the BMC blog http://t.co/dmuhQ7J #genomics #ISMB
Thu Jul 21 15:01:18 2011 bffo on way home 2 YYZ, leaving FRA on @aircanada, ~2 weeks on road. VIE for #ISMB and Kyoto (via KIX) 4 #ICGC. on plane feels like we r home
Thu Jul 21 13:54:11 2011 pjacock To all those who thanked me for #bosc2011 / #ISMB / #eccb2011 (live) tweets, no problem – it was an interesting experiment in note taking.
Thu Jul 21 12:36:39 2011 _lrr_ Simple yet very handy tool: fastapl (http://bit.ly/rhkKCl), had a poster at #ismb
Thu Jul 21 10:55:34 2011 druvus RT @dullhunk: Are videos of the #ISMB keynotes by Ashburner, Berger, Thornton, Serrano, Troyanskaya and Valencia online anywhere? #bioinformatics
Thu Jul 21 10:54:17 2011 druvus RT @GigaScience: Another applied example of #usegalaxy from Marc Bras, with a pipeline for SNP detection in the grapevine: MAPHiTS http://t.co/yVQ3POt #ISMB
Thu Jul 21 10:44:54 2011 bffo MT @GigaScience: #ISMB write-up in GigaBlog: Final impressions of Vienna: time 2 go w/t (work)flow… http://bit.ly/r6JaN7 #bioinformatics
Thu Jul 21 08:49:27 2011 fredebibs RT @druvus: ISMB/ECCB 2011 proceedings papers are online http://t.co/CkERlvb #ismb
Thu Jul 21 08:01:27 2011 iainh_z RT @GigaScience: Another #ISMB write-up in GigaBlog: Final impressions of Vienna: time to go with the (work)flow… http://t.co/ZVluBMa
Thu Jul 21 05:49:15 2011 heikkil RT @GigaScience: Another applied example of #usegalaxy from Marc Bras, with a pipeline for SNP detection in the grapevine: MAPHiTS http://t.co/yVQ3POt #ISMB
Thu Jul 21 03:47:52 2011 galaxyproject RT @GigaScience: Another applied example of #usegalaxy from Marc Bras, with a pipeline for SNP detection in the grapevine: MAPHiTS http://t.co/yVQ3POt #ISMB
Thu Jul 21 03:45:10 2011 galaxyproject RT @GigaScience: Another #ISMB write-up in GigaBlog: Final impressions of Vienna: time to go with the (work)flow… http://t.co/ZVluBMa
Thu Jul 21 02:43:42 2011 terryogara From chaos, beauty. RT @iGenomics: RW: Rather to fight noise, use noise to make a robust system. Add an oscillator to the system. #ismb
Wed Jul 20 22:03:00 2011 chlalanne RT @druvus: ISMB/ECCB 2011 proceedings papers are online http://t.co/CkERlvb #ismb
Wed Jul 20 21:30:05 2011 jxtx RT @GigaScience: Another #ISMB write-up in GigaBlog: Final impressions of Vienna: time to go with the (work)flow… http://t.co/ZVluBMa
Wed Jul 20 21:25:48 2011 GigaScience Another #ISMB write-up in GigaBlog: Final impressions of Vienna: time to go with the (work)flow… http://t.co/ZVluBMa
Wed Jul 20 20:47:27 2011 aadhyadi RT @ianholmes: "Don’t be afraid to work with people smarter than you. And work with people you like." Michael Ashburner, closing keynote, #ismb 2011
Wed Jul 20 20:40:34 2011 druvus ISMB/ECCB 2011 proceedings papers are online http://t.co/CkERlvb #ismb
Wed Jul 20 20:23:34 2011 olgabot @nerdgirls #ismb #ECCB11 was fantastic! Gained deep knowledge in my field, learned about new developments, and made some great friends
Wed Jul 20 20:23:19 2011 _lrr_ RT @RishaNarayan: Really enjoyed the ISMB/ECCB 2011 conference. Many thanks to the organisers. #ismb
Wed Jul 20 20:15:09 2011 lopantano RT @ianholmes: In closing #ismb keynote, Mike Ashburner quoted English philosopher(?) "Science is public knowledge. If it is not open, it is not science."
Wed Jul 20 19:59:57 2011 _lrr_ Had a great party at flex.at! RT @SaggiSardar: ISMB over for another year. Great conference. Now it’s time to enjoy Vienna! #ISMB
Wed Jul 20 18:39:15 2011 NatureRevGenet Congrats due to all the #ismb speakers for invariably clear and engaging talks
Wed Jul 20 16:46:36 2011 yeastgenome RT @bffo:Cherry,Ashburner and Blake; Sc, Dm & Mm, at the beginning of GO #ISMB http://yfrog.com/kl70850759j indeed a great closing lecture
Wed Jul 20 16:37:08 2011 dullhunk Are videos of the #ISMB keynotes by Ashburner, Berger, Thornton, Serrano, Troyanskaya and Valencia online anywhere? #bioinformatics
Wed Jul 20 16:00:19 2011 bffo Cherry, Ashburner and Blake; Sc, Dm & Mm, at the beginning of GO #ISMB http://yfrog.com/kl70850759j indeed a great closing lecture
Wed Jul 20 14:12:39 2011 tsucheta RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Wed Jul 20 14:00:21 2011 tsucheta RT @ianholmes: "Don’t be afraid to work with people smarter than you. And work with people you like." Michael Ashburner, closing keynote, #ismb 2011
Wed Jul 20 13:49:25 2011 avilella RT @ianholmes: Mike Ashburner claims "I never had to look for a job, and I’ve never been short of funding". Apparently 1970’s UK = science paradise #ismb
Wed Jul 20 12:52:39 2011 cbjones1943 RT @ianholmes: Once again I recommend Ashburner’s book "Won for All" for more such dirt-dishing details of millenial genomics http://t.co/otaIgvZ #ismb
Wed Jul 20 12:46:58 2011 genomeresearch RT @ianholmes: Once again I recommend Ashburner’s book "Won for All" for more such dirt-dishing details of millenial genomics http://t.co/otaIgvZ #ismb
Wed Jul 20 12:45:48 2011 cshperspectives Ashburner: Gerry Rubin "to his credit" recognized the merit of Venter’s approach but "Venter nearly screwed us" #ismb via @ianholmes
Wed Jul 20 12:21:24 2011 coding_doc RT @ianholmes: In closing #ismb keynote, Mike Ashburner quoted English philosopher(?) "Science is public knowledge. If it is not open, it is not science."
Wed Jul 20 12:17:39 2011 gthorisson Cannot believe my luck: flight from #ismb Vienna landed at Heathrhrow 25min early, so able to catch early bus to Oxford to meet colleagues
Wed Jul 20 11:47:34 2011 ClaireAinsworth RT @dgmacarthur: Check out @ianholmes for highlights of what sounds like a brilliant closing #ISMB keynote by Michael Ashburner.
Wed Jul 20 11:44:55 2011 sharmanedit RT @ianholmes: Once again I recommend Ashburner’s book "Won for All" for more such dirt-dishing details of millenial genomics http://t.co/otaIgvZ #ismb
Wed Jul 20 11:43:29 2011 sharmanedit Follow @ianholmes now for great live tweeting of Michael Ashburner’s fascinating closing keynote at #ismb 2011
Wed Jul 20 11:30:14 2011 ianholmes Once again I recommend Ashburner’s book "Won for All" for more such dirt-dishing details of millenial genomics http://t.co/otaIgvZ #ismb
Wed Jul 20 11:28:56 2011 ianholmes Ashburner gave credit to Suzi Lewis for the idea of 1999 Drosophila annotation jamboree & to Celera for funding it: "cheap consulting" #ismb
Wed Jul 20 11:27:59 2011 17thescorpion RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Wed Jul 20 11:27:20 2011 KatherineMejia RT @ianholmes: "Don’t be afraid to work with people smarter than you. And work with people you like." Michael Ashburner, closing keynote, #ismb 2011
Wed Jul 20 11:27:18 2011 ianholmes Ashburner’s early sequencing stories: getting Nature paper for ~5kb of fly DNA. Being scooped after talking about a gene to competitor #ismb
Wed Jul 20 11:24:58 2011 ianholmes Ashburner never intended to go into compbio. Even genetics was an accident. Recounted early adventures w/Phoenix, Vax, ARPAnet, gopher #ismb
Wed Jul 20 11:19:50 2011 dgmacarthur Check out @ianholmes for highlights of what sounds like a brilliant closing #ISMB keynote by Michael Ashburner.
Wed Jul 20 11:19:06 2011 gilbertjacka RT @ianholmes: Mike Ashburner claims "I never had to look for a job, and I’ve never been short of funding". Apparently 1970’s UK = science paradise #ismb
Wed Jul 20 11:18:10 2011 ianholmes Mike Ashburner claims "I never had to look for a job, and I’ve never been short of funding". Apparently 1970’s UK = science paradise #ismb
Wed Jul 20 11:17:08 2011 gawbul RT @ianholmes: Mike Ashburner noted that his own hacker, Aubrey de Grey, achieved "later notoriety as a gerontologist" #ismb
Wed Jul 20 11:16:10 2011 systemsbiology RT @ianholmes: Mike Ashburner turned down by Cambridge..for undergrad deg; took pleasure in turning down Zoology Dept chair 20yrs on #ismb
Wed Jul 20 11:15:23 2011 gawbul RT @ianholmes: Ashburner said Drosophila was his second love affair #ismb
Wed Jul 20 11:15:14 2011 gawbul RT @ianholmes: Ashburner accepted to do Genetics degree at Cambridge. Got married, learned genetics in a Naples lab that summer. Came back did degree #ismb
Wed Jul 20 11:14:05 2011 gawbul RT @ianholmes: Mike Ashburner was turned down by Cambridge Zoology for undergrad degree; took pleasure in turning down Zoology Dept chair 20yrs on #ismb
Wed Jul 20 11:12:58 2011 gawbul RT @ianholmes: In talking about Gene Ontology, Ashburner praised co-founders, as well as Barry Smith: "initially annoying" but "a true philosopher" #ismb
Wed Jul 20 11:12:54 2011 gawbul RT @ianholmes: "Don’t be afraid to work with people smarter than you. And work with people you like." Michael Ashburner, closing keynote, #ismb 2011
Wed Jul 20 11:12:51 2011 ianholmes Mike Ashburner noted that his own hacker, Aubrey de Grey, achieved "later notoriety as a gerontologist" #ismb
Wed Jul 20 11:12:27 2011 gawbul RT @ianholmes: In closing #ismb keynote, Mike Ashburner quoted English philosopher(?) "Science is public knowledge. If it is not open, it is not science."
Wed Jul 20 11:11:51 2011 ianholmes Ashburner’s #ismb pioneers: Roger Staden (Fred Sanger’s hacker), Doug Brutlag (Stanford compbio archive), Amos Bairoch (1st bio webserver)
Wed Jul 20 11:07:57 2011 ianholmes "Barry Smith – you either love him or you hate him" – Michael Ashburner, closing keynote, #ismb 2011
Wed Jul 20 11:06:55 2011 ianholmes "Don’t be afraid to work with people smarter than you. And work with people you like." Michael Ashburner, closing keynote, #ismb 2011
Wed Jul 20 11:05:44 2011 ianholmes In talking about Gene Ontology, Ashburner praised co-founders, as well as Barry Smith: "initially annoying" but "a true philosopher" #ismb
Wed Jul 20 11:04:44 2011 ilpuccio Reordering ideas after #BOSC2011 and #ISMB
Wed Jul 20 11:03:30 2011 ianholmes Ashburner clearly enjoyed his time at CalTech (think this was postdoc?) and mentioned Feynman – wonder if they met #ismb
Wed Jul 20 11:01:51 2011 ianholmes Ashburner said Drosophila was his second love affair #ismb
Wed Jul 20 11:01:27 2011 ianholmes Ashburner accepted to do Genetics degree at Cambridge. Got married, learned genetics in a Naples lab that summer. Came back did degree #ismb
Wed Jul 20 10:59:57 2011 ianholmes When Ashburner first invited to CalTech "I thought in my ignorance, why go to an Institute of Tech? Didn’t realize home of Drosophila" #ismb
Wed Jul 20 10:58:42 2011 ianholmes Gerry Rubin "to his great credit" recognized merit of Craig Venter’s approach to sequencing. But "Venter nearly screwed us" -Ashburner #ismb
Wed Jul 20 10:56:57 2011 ianholmes Mike Ashburner was turned down by Cambridge Zoology for undergrad degree; took pleasure in turning down Zoology Dept chair 20yrs on #ismb
Wed Jul 20 10:56:18 2011 wimufi RT @ianholmes: In closing #ismb keynote, Mike Ashburner quoted English philosopher(?) "Science is public knowledge. If it is not open, it is not science."
Wed Jul 20 10:53:13 2011 ianholmes Mike Ashburner’s #ismb keynote concluded with "lessons". It’s all about luck. But must exploit that luck. Get a guru/mentor/parent. Be open.
Wed Jul 20 10:51:18 2011 ianholmes In fine (blissfully un-PC) form, Mike Ashburner also relished stories of competition with "the Germans", "the Spanish" etc. #ismb
Wed Jul 20 10:50:23 2011 ianholmes Ashburner also noted (and NB I’m not endorsing every quote! but his #ismb keynote WAS juicy) that he’d never be an American citizen.
Wed Jul 20 10:49:05 2011 ianholmes Mike Ashburner was offered a UK biowar job out of college, but refused. Wonders now if he would’ve got security clearance. Hopes not. #ismb
Wed Jul 20 10:47:53 2011 ianholmes In closing #ismb keynote, Mike Ashburner quoted English philosopher(?) "Science is public knowledge. If it is not open, it is not science."
Wed Jul 20 10:45:13 2011 ianholmes To say Gene Ontology received coolly at #ismb Halkidiki in ’97 an understatement; Ashburner "almost laughed out of room" by Sander, Durbin
Wed Jul 20 10:43:23 2011 ianholmes I highly recommend Ashburner’s book "Won for All". Candid autobiography of the millenial corporate genome wars http://t.co/otaIgvZ #ismb #fb
Wed Jul 20 10:43:10 2011 druvus I have enjoyed following #ismb tweets from my sunny garden
Wed Jul 20 10:40:34 2011 ianholmes I *really* enjoyed Michael Ashburner’s closing #ISMB keynote. I was unable to tweet at the time but will record some highlights.
Wed Jul 20 10:40:11 2011 ianholmes Nice –> @nutrigenomics @iGenomics Ron Weiss: Don’t fight noise; use it to make gene circuits robust. Add an oscillator to the system. #ismb
Wed Jul 20 10:33:54 2011 ianholmes RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Wed Jul 20 10:33:51 2011 ianholmes RT @iGenomics: Michael Ashburner: Science is public knowledge. If it is not open, it is not science. #ismb
Wed Jul 20 08:59:25 2011 samuellampa RT @iGenomics: A Bioinformatics training resource: http://www.biotnet.org/ #ismb
Wed Jul 20 08:37:30 2011 biomol_info RT @iGenomics: A Bioinformatics training resource: http://www.biotnet.org/ #ismb
Wed Jul 20 08:25:30 2011 gthorisson On my way home from #ISMB. Vienna airport is crowded, stuffy and not very pleasant. But, it has free wireless Internet so it ROX.
Wed Jul 20 06:56:48 2011 bffo block dates for #ISMB next year (and surrounding dates for SIGs): July 15-17 2012 , this year was great, c u all in LA next year!
Wed Jul 20 06:28:06 2011 iliasSkalk RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Wed Jul 20 06:27:46 2011 iliasSkalk RT @iGenomics: Michael Ashburner: Science is public knowledge. If it is not open, it is not science. #ismb
Wed Jul 20 01:33:31 2011 yeastgenome RT @gthorisson: Fantastic #ISMB keynote from Michael Ashburner. What an inspiration. Key msgs: ‘collaborate with people brighter than you’ and ‘openness’
Wed Jul 20 01:32:27 2011 yeastgenome RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Wed Jul 20 00:00:26 2011 robymuhamad RT @timjph: @iGenomics here’s report I mentioned: 3.8b$ on human genome project; 310k jobs created & ~800b$ economic impact http://j.mp/rbXSbk #ismb
Tue Jul 19 23:53:57 2011 robaganrab RT @iGenomics: Michael Ashburner: Chance happens in life. You have to exploit it #ismb
Tue Jul 19 23:38:41 2011 scroeser RT @_lrr_: Michael Ashburner: "If knowledge is not public it is not science. Period." #ismb #eccb11 #openscience
Tue Jul 19 22:53:00 2011 ematsen RT @iddux: HumanN, a new metagenomic analysis package from Huttenhower http://j.mp/pHkWhk #ISMB
Tue Jul 19 22:36:32 2011 SaggiSardar ISMB over for another year. Great conference. Now it’s time to enjoy Vienna! #ISMB
Tue Jul 19 22:30:06 2011 aaronquinlan RT @iGenomics: Michael Ashburner: Follow Syndey Brenner, he will not use slides because a good phrase is worth thousand of Powerpoint slides. #ismb
Tue Jul 19 21:09:21 2011 aadhyadi RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Tue Jul 19 21:09:00 2011 chofski RT @SUPERFAMILY: Congratulations to Adam Sarder on winning the outstanding poster award at #ISMB (it’s poster F22 if you want to see it!)
Tue Jul 19 20:42:57 2011 wspoonr RT @pjacock: Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openscience
Tue Jul 19 19:36:10 2011 simon_andrews After all of the #ISMB delegates have left, our hotel wifi mysteriously starts working again.
Tue Jul 19 19:14:33 2011 KamounLab RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Tue Jul 19 19:14:24 2011 KamounLab RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Tue Jul 19 18:57:14 2011 julientap RT @iddux: HumanN, a new metagenomic analysis package from Huttenhower http://j.mp/pHkWhk #ISMB
Tue Jul 19 18:49:04 2011 alalejandro RT @iGenomics: Michael Ashburner: Science is public knowledge. If it is not open, it is not science. #ismb
Tue Jul 19 18:48:56 2011 alalejandro RT @gthorisson: Fantastic #ISMB keynote from Michael Ashburner. What an inspiration. Key msgs: ‘collaborate with people brighter than you’ and ‘openness’
Tue Jul 19 18:48:07 2011 alalejandro If only people would understand how essential this is! MT @keesvanbochove Ashburner #ismb: don’t be proprietary abt your research, be open.
Tue Jul 19 18:46:17 2011 RishaNarayan Really enjoyed the ISMB/ECCB 2011 conference. Many thanks to the organisers. #ismb
Tue Jul 19 18:42:56 2011 caseybergman RT @gthorisson: Fantastic #ISMB keynote from Michael Ashburner. What an inspiration. Key msgs: ‘collaborate with people brighter than you’ and ‘openness’
Tue Jul 19 18:40:34 2011 bergmanlab RT @olgabot: #ismb "If knowledge is not public it is not science" – Michael Ashburner
Tue Jul 19 18:40:33 2011 caseybergman RT @olgabot: #ismb "If knowledge is not public it is not science" – Michael Ashburner
Tue Jul 19 18:31:02 2011 iddux HumanN, a new metagenomic analysis package from Huttenhower http://j.mp/pHkWhk #ISMB
Tue Jul 19 18:19:10 2011 iddux @_lrr_: @fionabrinkman learned at #ismb that what we call "analogs" today used to be called "orthologs" by 1s… (cont) http://deck.ly/~yFYyV
Tue Jul 19 18:18:07 2011 iddux @_lrr_: @fionabrinkman learned at #ismb that what we call "orthologs" today used to be called "analogs" by 1st evolutionary scientists.
Tue Jul 19 17:27:04 2011 4a6a5a RT @simon_andrews: 2 people have now confused me with Simon Anders (DeSeq etc) I should learn more about RNASeq to be able to bluff better. #ismb
Tue Jul 19 17:24:43 2011 Accelrys RT @Fabio11211 #ISMB conf draws to an end! Strong presence & interest at Accelrys booth for NextGenSeq & Protein Modelling solutions
Tue Jul 19 17:19:48 2011 Chris_Evelo MT @pjacock: Gary Bader suggested http://bit.ly/mPS9LS for shared #bioinformatics tutorials, eg has introduction Cytoscape #ISMB Workshop
Tue Jul 19 17:17:55 2011 suknamgoong RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Tue Jul 19 17:16:43 2011 cytoscape RT @pjacock: Gary Bader suggested http://t.co/jNFVbjr as one project to shared #bioinformatics tutorials, eg has introduction Cytoscape #ISMB Workshop
Tue Jul 19 17:02:34 2011 BetaScience @bgood Wish I was at #ismb with you. I have some great memories from that conference and #Vienna. #pre-kids
Tue Jul 19 16:55:50 2011 betsyrolland RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Tue Jul 19 16:50:41 2011 jpatanooga RT @keesvanbochove: Closing remark of Michael Ashburner, a godfather of #bioinformatics, at #ismb: don’t be proprietary about your research, be open.
Tue Jul 19 16:22:44 2011 gailfdavies RT @olgabot: #ismb "If knowledge is not public it is not science" – Michael Ashburner
Tue Jul 19 16:20:55 2011 iGenomics @westr Yes, indeed. Michael Ashburner borrowed it to tell 3 life lessons. #ismb
Tue Jul 19 16:18:38 2011 pierrepo RT @gthorisson: Fantastic #ISMB keynote from Michael Ashburner. What an inspiration. Key msgs: ‘collaborate with people brighter than you’ and ‘openness’
Tue Jul 19 16:12:37 2011 FigShare RT @olgabot: #ismb "If knowledge is not public it is not science" – Michael Ashburner
Tue Jul 19 16:12:19 2011 tweetruth RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Tue Jul 19 16:11:49 2011 FigShare RT @keesvanbochove: Closing remark of Michael Ashburner, a godfather of #bioinformatics, at #ismb: don’t be proprietary about your research, be open.
Tue Jul 19 16:11:22 2011 kshameer RT @antidemagogue: MA: Knowledge which is not public is not science, period. #ismb #bioinformatics #genomics
Tue Jul 19 16:11:08 2011 iscbsc for those student in Vienna, at 8 at stephanplatz #scs2011 #ISMB
Tue Jul 19 16:11:04 2011 kshameer RT @keesvanbochove: Closing remark of Michael Ashburner, a godfather of #bioinformatics, at #ismb: don’t be proprietary about your research, be open.
Tue Jul 19 16:04:14 2011 c_rdzl RT @iGenomics: Michael Ashburner: Science is public knowledge. If it is not open, it is not science. #ismb
Tue Jul 19 16:03:00 2011 ffalconi RT @iGenomics: Michael Ashburner: Chance happens in life. You have to exploit it #ismb
Tue Jul 19 16:01:34 2011 science3point0 RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Tue Jul 19 16:01:01 2011 GigaScience RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Tue Jul 19 15:59:27 2011 pjacock For 2012, #ISMB in Long Beach, USA, and #ECCB in Basel, Switzerland
Tue Jul 19 15:59:21 2011 keesvanbochove RT @timjph: @iGenomics here’s report I mentioned: 3.8b$ on human genome project; 310k jobs created & ~800b$ economic impact http://j.mp/rbXSbk #ismb
Tue Jul 19 15:59:20 2011 fredebibs RT @gthorisson: Fantastic #ISMB keynote from Michael Ashburner. What an inspiration. Key msgs: ‘collaborate with people brighter than you’ and ‘openness’
Tue Jul 19 15:55:20 2011 pjacock RT @gthorisson: Fantastic #ISMB keynote from Michael Ashburner. What an inspiration. Key msgs: ‘collaborate with people brighter than you’ and ‘openness’
Tue Jul 19 15:54:42 2011 keesvanbochove Closing remark of Michael Ashburner, a godfather of #bioinformatics, at #ismb: don’t be proprietary about your research, be open.
Tue Jul 19 15:54:34 2011 pjacock Embarrassingly many of the #ISMB / #eccb2011 prize winners haven’t stayed for the prize announcements in the closing session
Tue Jul 19 15:52:34 2011 _lrr_ Michael Ashburner: "If knowledge is not public it is not science. Period." #ismb #eccb11 #openscience
Tue Jul 19 15:52:31 2011 gthorisson RT @antidemagogue: MA: Knowledge which is not public is not science, period. #ismb
Tue Jul 19 15:52:26 2011 ajordens RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Tue Jul 19 15:51:54 2011 gthorisson Fantastic #ISMB keynote from Michael Ashburner. What an inspiration. Key msgs: ‘collaborate with people brighter than you’ and ‘openness’
Tue Jul 19 15:49:33 2011 pjacock RT @iGenomics: Michael Ashburner: Chance happens in life. You have to exploit it #ismb
Tue Jul 19 15:49:30 2011 pjacock RT @iGenomics: Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Tue Jul 19 15:48:53 2011 pjacock #ISMB #killerapp award goes to Syed Asad Rahman for EC-BLAST which searches for similar enzymes based on chemical knowledge
Tue Jul 19 15:48:08 2011 antidemagogue MA: Knowledge which is not public is not science, period. #ismb
Tue Jul 19 15:48:02 2011 iGenomics Michael Ashburner: Chance happens in life. You have to exploit it #ismb
Tue Jul 19 15:46:51 2011 pjacock RT @iGenomics: Michael Ashburner: Science is public knowledge. If it is not open, it is not science. #ismb
Tue Jul 19 15:46:35 2011 druvus RT @pjacock: Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Tue Jul 19 15:46:34 2011 antidemagogue MA: Exploit your luck. Collaborate. Communicate research openly. #ismb
Tue Jul 19 15:46:21 2011 iGenomics Michael Ashburner: Try to work with people who are smarter than you are. #ismb
Tue Jul 19 15:45:45 2011 pjacock Michael Ashburner #ISMB tells young scientists to be open in their work. If knowledge is not open, it is not science! #openaccess #opendata
Tue Jul 19 15:45:37 2011 olgabot #ismb "If knowledge is not public it is not science" – Michael Ashburner
Tue Jul 19 15:45:17 2011 iGenomics Michael Ashburner: Science is public knowledge. If it is not open, it is not science. #ismb
Tue Jul 19 15:44:55 2011 biobrichter #ISMB ashburner talk. informative, brings back the memories of the seq 90’s, but very ashburner-centric. True:collab with brighter people!
Tue Jul 19 15:44:26 2011 pjacock Michael Ashburner wrapping up his #ISMB keynote, describes career as a random walk. Recommends collaborating with people brighter than you!
>>>
Balkan Beat Box, with some good advice for me, and for anyone else with a grant proposal deadline tomorrow:



















