The art of motivating employees. Interesting insights and beautiful illustrations. Also, a good mention of the open source and collaborative content movements.
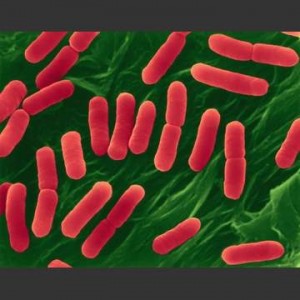
![]() In case you have been vacationing in a parallel universe in the past two days, you should have heard about the new synthetic bacterium created at the J Craig Venter Institute. In a nutshell, the scientific team synthesized an artificial chromosome of the bacterium Mycoplasma mycoides and transferred it to another bacterium, Mycoplasma capricolum. The capricolum cells with the mycoides genome proved viable, and were named Mycoplasma mycoides JCVI-syn1.0. Even more briefly they synthesized Bug A’s DNA from scratch, put it in bug B, turning B into A.
In case you have been vacationing in a parallel universe in the past two days, you should have heard about the new synthetic bacterium created at the J Craig Venter Institute. In a nutshell, the scientific team synthesized an artificial chromosome of the bacterium Mycoplasma mycoides and transferred it to another bacterium, Mycoplasma capricolum. The capricolum cells with the mycoides genome proved viable, and were named Mycoplasma mycoides JCVI-syn1.0. Even more briefly they synthesized Bug A’s DNA from scratch, put it in bug B, turning B into A.
I wanted to write a blog post about it. I really did. Something original, inspiring, funny, critical and deep. But so many others beat me to it, so no matter what angle I took, it’s already been covered in the last 24 hours. Informative? Yes. Debateable achievement? Yes yes, and yes. Thoughts from bigshots? Yes. Funny? totally. Religiously suspect? Verily. Government weighing in? Naturally. Reddit? Yes, even Reddit! (Thanks Shirley!)
So here’s the interview Science journal conducted with Craig Venter:
Or, if you’d rather, the Scorpions’ comment:
And the paper in Science. I’m done.
Gibson, D., Glass, J., Lartigue, C., Noskov, V., Chuang, R., Algire, M., Benders, G., Montague, M., Ma, L., Moodie, M., Merryman, C., Vashee, S., Krishnakumar, R., Assad-Garcia, N., Andrews-Pfannkoch, C., Denisova, E., Young, L., Qi, Z., Segall-Shapiro, T., Calvey, C., Parmar, P., Hutchison, C., Smith, H., & Venter, J. (2010). Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome Science DOI: 10.1126/science.1190719
Big party at Science journal today, with the publication of a comprehensive draft Neanderthal genome. (Free access, nice going Science). Actually, it is a partially assembled draft of 60% of the total genome, but 60% of the genome from a human that was last seen on Earth 28,000 years ago is quite an achievement. The DNA was extracted from three bone fragments found in a cave in Croatia. The bones came from different individuals, so it is a composite rather than the draft genome of a single person. Also, the coverage — number of times each base was sequenced on average — is only 1.3, which is quite low,which means that in all probability there are still quite a few sequencing errors. Sequencing errors get weeded out as more overlapping regions get sequenced: the spurious sequencing errors get diluted with every new sequencing run. Still, we can learn many things about ourselves and our recently-extinct human companions even from a partial, fragmented and somewhat still-erroneous sequence.
![]()
Until now we had only three good, full copies of Neanderthal mitochondrial DNA. Mitochondrial DNA codes for a relatively small number of genes, and is inherited through the mother. There are regions on the DNA that are more variable, which helped us map the genetic distance between modern humans and Neanderthals. Actually, I did that in my bioinformatics course this year. The conclusion reached by the researchers who did that, as well as by my students was that modern humans are much closer to each other than to Neanderthals, and that the genetic distance is large enough to make humans and Neanderthals two different species.
But the genomic DNA reveals that between 1-4% of modern human DNA is close enough to equivalent segments from Neanderthal DNA to claim a gene flow and hence interbreeding. Second, Neanderthals are closer to European and Asian humans than they are to those from sub-Saharan Africa. Actually, modern humans from sub-Saharan Africa do not have the Neanderthal DNA markers. This fits well with the current hypothesis on the origins of Neanderthals: they were the descendants of the first Out of Africa migration, some 500,000 years ago. They lived in Europe and Asia, while the ancestors of modern humans lived in Africa. The second out of Africa migration happened about 100,000 years ago, which is when modern humans first encountered Neanderthals and, as shown now by this study, have interbred with them.
On the spelling of Neandert(h)al: you many have noticed that the spelling in the Science paper, and in many other places is “Neandertal”, whereas others spell it as “Neanderthal”. (My Firefox spellchecker actually does not like the h-less version.) Talkorigins.org explains why that is.
Now, if you will kindly excuse me, I have some slides and a student lab to revise for next year.
Green, R., Krause, J., Briggs, A., Maricic, T., Stenzel, U., Kircher, M., Patterson, N., Li, H., Zhai, W., Fritz, M., Hansen, N., Durand, E., Malaspinas, A., Jensen, J., Marques-Bonet, T., Alkan, C., Prufer, K., Meyer, M., Burbano, H., Good, J., Schultz, R., Aximu-Petri, A., Butthof, A., Hober, B., Hoffner, B., Siegemund, M., Weihmann, A., Nusbaum, C., Lander, E., Russ, C., Novod, N., Affourtit, J., Egholm, M., Verna, C., Rudan, P., Brajkovic, D., Kucan, Z., Gusic, I., Doronichev, V., Golovanova, L., Lalueza-Fox, C., de la Rasilla, M., Fortea, J., Rosas, A., Schmitz, R., Johnson, P., Eichler, E., Falush, D., Birney, E., Mullikin, J., Slatkin, M., Nielsen, R., Kelso, J., Lachmann, M., Reich, D., & Paabo, S. (2010). A Draft Sequence of the Neandertal Genome Science, 328 (5979), 710-722 DOI: 10.1126/science.1188021
No, not the flesh-blood-and-feathers penguin, but rather Tux, the beloved mascot of the Linux operating system. Compared with Escherichia coli, the model organism of choice for microbiologists.
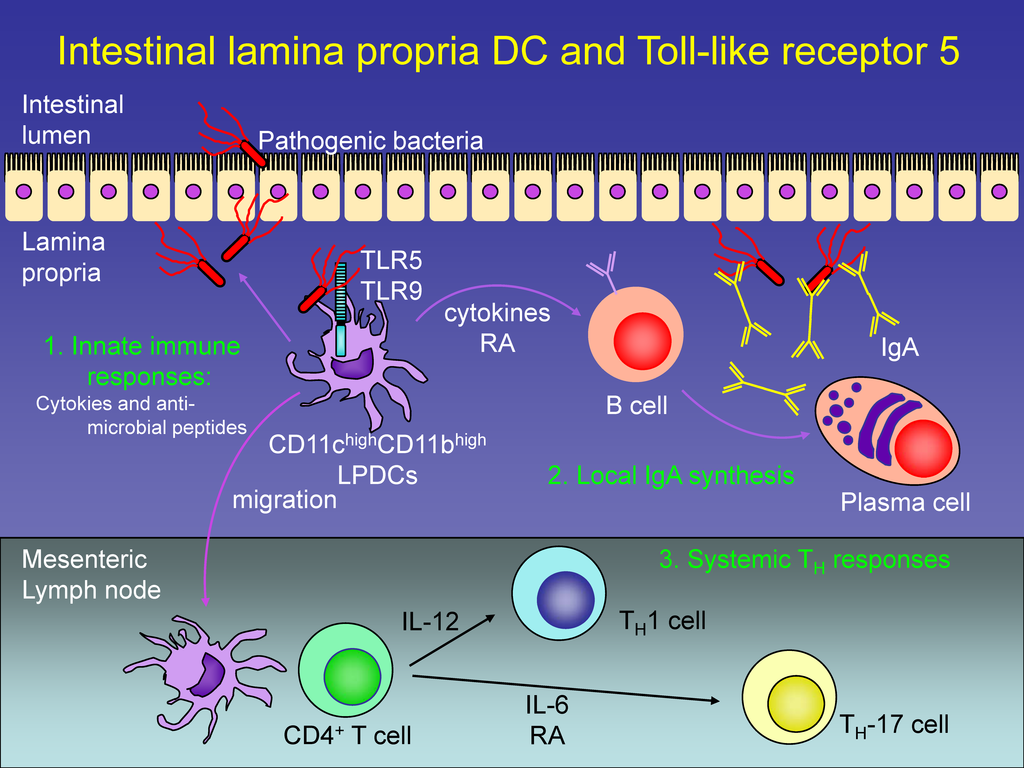
We refer to DNA as “the book of life”; some geeks refer to it as the “operating system of life”. Just like in a computer’s operating system, DNA contains all the instructions on how to “execute” life and to keep things humming. Many genes make proteins or RNA than act as switches to activate the synthesis of other proteins, sometimes in a two- three- or higher level hierarchy. These switches are conditional, based on environmental conditions, or whether it’s time to replicate the DNA and divide into two daughter cells, and so on. Some genes activate the transcription of other genes, but are not regulated themselves by other genes, those can be dubbed “master regulators”. Some genes are both activated by other genes, and activate other genes themselves: “middle management”. Finally, there are genes that are activated, but do not regulate other genes: the “workhorses”. This information, known as the transcriptional regulatory network exists for 1,378 genes of the E. coli bacterium.
![]()
Paralleling this in Linux, there are programs that call other programs; again, in a hierarchical fashion. According to the calling structure, they also can be dubbed Master Regulators (calling other programs but not being called themselves), Middle Management (calling other programs and being called), and Workhorse (only being called).
Koon-Kiu Yan and his colleagues from Yale mapped the program call graph in Linux by setting each program as a node and drawing lines to the programs that call it, and to the programs it calls. They did the same thing for E. coli‘s transcriptional regulatory network. Here are the graphs they got:
So it seems like Linux is middle-management heavy, whereas E. coli is workhorse heavy. 30% of Linux programs are top management, as opposed to only 5% in E. coli.
Looking at the actual functions for the genes/programs, it seems that Linux programs also have much more of a functional redundancy than in E. coli: 3.5% of E. coli‘s genes have “reusable” functions, as opposed to 8.4% of Linux programs. But if we look at entire working subgraphs of these two graphs, the subgraph overlap in Linux is 87%, whereas in E.coli the overlap is only 4.3%. This means that the division of labor in E. coli is much more distinct than in Linux. There are many ways of activating the same hierarchy in Linux, but in E. coli there is rarely more than one way to do it. Note that Linux is top-heavy, whereas E. coli has a pyramid-like structure. It is pretty obvious that the Workhorse modules in Linux go through heavy reuse while those in E. coli do not.
The scientists then decided to look into how these two networks developed. The oldest genes in E. coli are the Workhorses, whereas the regulatory genes in middle and top management arrived more recently. In contrast, the newest programs — the most heavily rewritten ones– in Linux are the Workhorses, whereas the ones in the management echelons are less changed than their predecessors. The oldest programs are those that are in Middle Management. they are also the most abundant type in Linux’s call graph.
Who are the Workhorses in E. coli? Those are mostly enzymes, the proteins that catalyze specific biochemical functions. As a rule, enzymes are very specific: an enzyme would catalyze only one type of reaction, and only with a very specific chemical (substrate). Examples are enzymes that break up sugars: there is a specific enzyme for every type of sugar molecule. Who are the Workhorses in Linux? Those are the functions that get used all the time in thousands of different programs: strlen (measuring a character string’s length) or malloc (allocating memory for a data structure). The Workhorses in Linux are non-specific while the Workhorses in E. coli are very specific.
So how to account for these differences? Nothing in biology makes sense except in light of evolution, and we have to look to the evolutionary history of both the bacterial and the computational systems for answers. The major constraint in E. coli‘s evolution is fitness. If something breaks down in E. coli‘s Workhorse it wont get passed on to the next generation: the cell with the lethal mutation would never reproduce and will get thrown into Darwin’s rubbish bin. This leads to single-function workhorses because a multi-functional Workhorse would be too prone to messing too many systems up when it mutates, and would never make it to the next generation, which is why the Workhorses in E. coli‘s call graph have a lower connectivity that those in Linux’s call graph.
The authors conclude that the E. coli‘s call graph evolved bottom-up, with system robustness being the main selective trait. In contrast, Linux evolved top-bottom, with reusability of the Workhorses being the main selective trait. Reusability and robustness are tradeoffs. In the case of a man-made system like Linux, bugs in reusable modules are is not a problem, since Workhorse bugs are easily fixed in the next release. It is much less costly, in coding time, to tweak existing Workhorses than to build new ones. Mutations in reusable workhorses in E. coli would weed out those kinds of proteins from the gene pool, and therefore E. coli‘s Workhorses are not reusable.
I’m not exactly sure what insight we can get by comparing natural vs. man-made networks. But hey, sometimes science is not about insight – sometimes is just about being totally cool; and The Coolness is strong with this work.
Yan, K., Fang, G., Bhardwaj, N., Alexander, R., & Gerstein, M. (2010). Comparing genomes to computer operating systems in terms of the topology and evolution of their regulatory control networks Proceedings of the National Academy of Sciences DOI: 10.1073/pnas.0914771107
Combrex is an exciting new project at Boston University to bridge computational and experimental techniques to functionally annotate proteins. They are hiring, see below:
JOB POST
We are seeking to hire a creative computational scientist for a
transformative project: COMBREX: A Computational Bridge to Experiments.The work will involve building a novel resource that combines databases,
science, social networking and machine learning.The position is available immediately.
For some preliminary information pls. see
BS or MS in Computer Science, Informatics, Engineering or related field is required.
Applicants with PhD’s would be considered for a separate Research Associate position.
Pls send CV and names (emails) of two references to:
Prof. Simon Kasif
Subject Line: COMBREX POSITION
Nature is colorful. And the family of pigments that is mostly responsible for these colors are carotenoids. Carotenoids make the apples and tomatoes red, the lemons and grapefruit yellow, the pumpkins oranges and, yes carrots, (from which their name is derived), orange.
Carotenoids also make flamingos and salmon pink, and color the puffin’s bill orange. But those animals cannot produce carotenoids: rather carotenoids are in their diet, and in the case of flamingos and puffins they have a physiological mechanism of concentrating the carotene molecules and bringing them to display their strong colors. Indeed Some of us also use carotenoids as an ornamental physiological addition: remember that horrid orange tan Auntie Mae sported last time you saw her in the dead of winter? Beta-carotene pills. Carotene is not just ornamental in animals, they are also important for eyesight, the immune system, and in decreasing DNA damage that may lead to cancer.
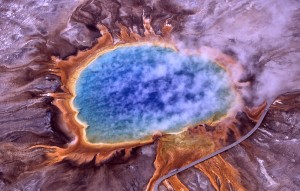
The orange ring surrounding Grand Prismatic Spring is due to carotenoid molecules, produced by huge mats of algae and bacteria. Source: wikimedia commons. Credit: Jim Peaco, US National Park Service
An article published today in Science shows the first case of animals synthesizing carotenoids. Nancy Moran and Tyler Jarvik form the University of Arizona looked at the recently sequenced genome of the pea aphid. The pea aphid is known for having two different colors: green and red. It was not very clear though how the aphids got their color. Aphids feed on sap, and sap does not contain carotenoids. When looking at the genomes of the aphids, Moran and Jarvik found that they contained genes for synthesizing carotenoids: this is the first time carotenoid synthesizing genes are found in animals. The question they naturally asked is “where did those genes come from”? The animal kingdom does not contain genes for making carotenoids, so how come aphids have them? Indeed, when they looked for the most similar genes to the aphid carotenoid synthesizing genes they found that they came from fungi, which means they somehow jumped between fungi and aphids, in a process known as horizontal gene transfer. Horizontal gene transfer is not unheard-of in animals, and is actually quite common in plants (yeah, fungi are not plants, I know that), but this is the first time someone has shown a jump from fungi to animals, and that the trait that this gene conveys — color — became embedded and functional in the genome.
Aphid color is important: red aphids get picked easily by predators off green plants, and vice-versa. So there is an evolutionary aspect here: the carotenoid genes play a role in the predator-driven selection of aphids. So in the case of aphids, as opposed to puffins and flamingo, the selective pressure is that of predation, not of mating. (I’ll refrain from comments about Auntie Mae.)
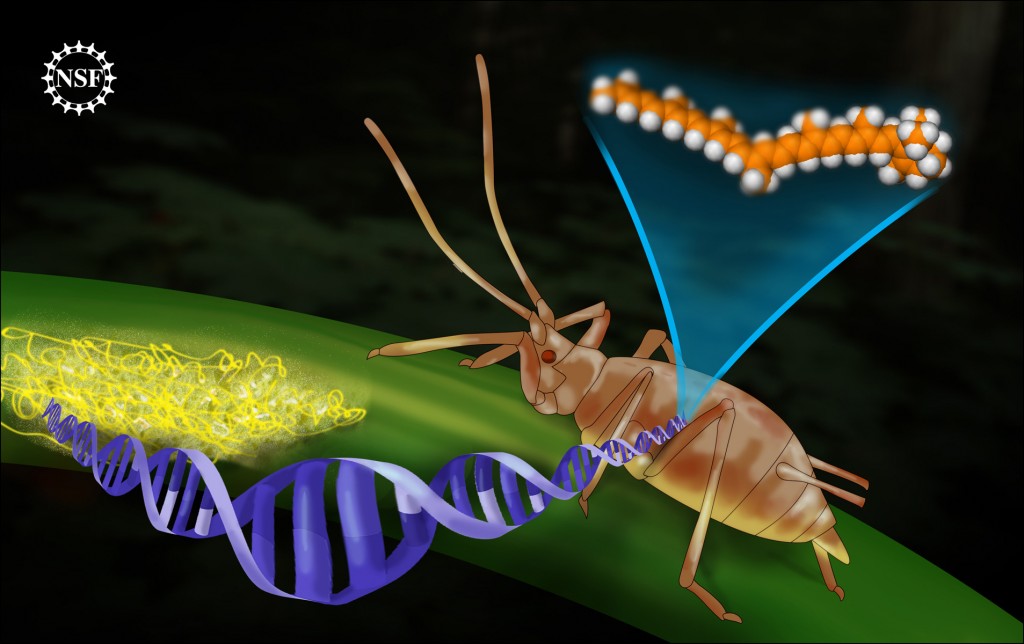
"Long ago, an ancestor to today's pea aphid somehow internalized a large important chunk of DNA from a fungus. This DNA now allows the aphid to generate its own carotenoid molecules. All animals need carotenoids for body functions as important as eyesight. However this aphid is the only organism in the Animal Kingdom so far to have been reported capable of producing it internally. The rest of us must forage for foods such as carrots, containing carotenoids. The precise way the DNA transfer occurred is not yet understood; however patterns within the DNA conclusively show a link to a fungus. DNA transfer from fungus to animal is unprecedented." (text taken from the NSF announcement). Credit: Zina Deretsky, National Science Foundation
As an aside, many of our pseudogenes and other contents of “junk DNA” are thought to have been acquired by horizontal gene transfer. Still, this is the first time a case of gene transfer that is so clear between two different kingdoms. However, I have the sneaking suspicion that as we sequence more animal, plant, fungal and other genomes of multicellular organism, we would find more cases of “large-leap” HGT of functional genes happening: we just don’t have enough genomes yet to appreciate the frequency of these occurrences!
 Update: this post has been slashdotted. Exercise extreme caution.
Update: this post has been slashdotted. Exercise extreme caution.
Update II: this post has been submitted for the NESCent travel award for the Science Online 2011 conference.
Moran, N., & Jarvik, T. (2010). Lateral Transfer of Genes from Fungi Underlies Carotenoid Production in Aphids Science, 328 (5978), 624-627 DOI: 10.1126/science.1187113
Beautiful video showing the mathematical beauty of nature, or the natural beauty of math.
Here’s what I managed to figure out:
0:08-0:44 – Fibonacci sequence
0:45-1:40 – The Golden Ratio
2:40 – Delauney triangulations leading to Voronoi diagrams (2:56 and to the end)
Would you have your genome sequenced for free? Conditions: you must license it for all use; a liberal CC-no attribution-like license which allows for commercial use as well. Also, your genome will be made public with many personal data such as age, height, sex, weight, ethnicity, personal status (we want to find the “money making gene” and the “fecundity gene”) medical history, some family history but not name or exact geographic location. Note that these metadata may make you traceable, although not easily. To fully appreciate the genotype-phenotype connection, your doctor will upload your medical records with each visit & checkup. Hey, once you contract that late-onset Cheetos addiction at the age of 65, we want to know about it. Hmmm, not so free after all, maybe. Still you may want to do it. Tick in your answer in the blue box in the right sidebar. HT to Mickey for the idea.
For old-school geeks who spent the better time of their childhood prefecting their Space Invaders and Donkey Kong skills. A NYC disaster movie meets… well, something. Hat tip to Mickey.
Recently, a judge in Federal District Court in Manhattan ruled that Myriad’s patents on BRCA1 and BRCA2 genes were invalid, being a “products of the law nature” and could be patented no more than, say, mount Everest. These two genes are associated with breast and ovarian cancer, and are used in testing for susceptibility to these types of cancer — and for the patent’s duration, using Myriad’s labs. The ruling, if it holds up in appeals, will change the way pharmaceutical business is done: there are over 4,300 gene patents today. BRCA2 tests cost $3,000 in the US, where Myriad has exclusivity. In some provinces of Canada, where Myriad’s exclusivity is not honored, BRCA tests cost considerably less. As an aside, one of the successes that the plaintiffs attribute to the verdict is the contribution to women’s health. True, but not exclusively so: there is growing evidence that BRCA1/2 mutations are associated with pancreatic cancer and testicular cancer.
Stephen Colbert has something to say about it; but in this case, although he is his usual facetiously hilarious self, he seemed to confuse the ACLU, who was one of the plaintiffs. Actually, he confused me too. His arguments for the patent invalidation seem a tad self-defeating, rather unusual for Colbert. See for yourself.
| The Colbert Report | Mon – Thurs 11:30pm / 10:30c | |||
| Formula 01 Liquid Genetic Material | ||||
|
||||
AMOS is a suite of genome assembly and editing software. It includes assemblers, validation, visualization, and scaffolding tools. I have been having some issues installing AMOS on Ubuntu 9.10. Specifically, Ubuntu 9.10 has gcc 4.4, which breaks the compilation of the AMOS release version. However, the development version has been fixed to accommodate that.
If you don’t know which Ubuntu version you are running, type:
$ lsb_release -a
No more than fifteen minutes after I posted my Q to the amos-help mailing list, Florent Angly came through with a solution. I am posting his email here.
Hi,
This issue was fixed in the development version of AMOS. See below for instructions on how to install this version on Ubuntu:
Download either the regular or development version of AMOS. As of April 4, 2010,
Minimo is only available from the development version of AMOS.
i/ The regular AMOS version is available from http://sourceforge.net/projects/amos/files/, e.g.:
$ wget http://sourceforge.net/projects/amos/files/amos/2.0.8/amos-2.0.8.tar.gz/download
ii/ The development version of AMOS is in a CVS repository. To get it, run:
$ cvs -z3 -d:pserver:anonymous@amos.cvs.sourceforge.net:/cvsroot/amos co -P AMOSIn the directory where the AMOS file are located, run the following to install
the prerequisites:
$ sudo aptitude install ash coreutils gawk gcc automake mummer mummer-doc libboost-devFor the Hawkeye component of AMOS, you need Qt3:
$ sudo aptitude install libqt3-headersFor the standard version of AMOS, skip to next step, but for the CVS development version, first, run:
$ ./bootstrapThen regardless of the version:
$ ./configure –with-Qt-dir=/usr/share/qt3 –prefix=/usr/local/AMOS
$ make
$ make check
$ sudo make install
$ sudo ln -s /usr/local/AMOS/bin/* /usr/local/bin/Now all the programs shipped in AMOS should be available from the command-line.
For example try:
$ Minimo -h
Regards,Florent
You will need the AMOS development version for Ubuntu 9.10 (and above, presumably), but the regular version for 9.04 (and below). If you are getting the development version, you will also need to install cvs on your machine:
$ sudo aptitude install cvs
Hope this helps anyone struggling with installing AMOS on Ubuntu or other Linux platforms.
It seems like there is no institution that is more criticized in science than that of the peer-review system — an no one that is less mutable. While published paper evaluation metrics are being revised (such as the recently introduced PLoS article level metrics, or the Australian National Health and Medical Research Council abandonment of the Thomson Reuters impact factor system), the peer review system seems like it is here to stay. When asked, most scientists would probably paraphrase Churchill: “peer review is the worst system for judging science, except all others that have been tried from time to time”. (However, Churchill did have other working state models to compare with Democracy, whereas peer-review seems to have no, um, peers.) The latest diagnostic comes from Errol Friedberg (no relation to me), editor in chief of DNA Repair.
“—if peer review is so central to the process by which scientific knowledge becomes canonized, it is ironic that science has little to say about whether it works.” — J.P. Kassirer and E.W. Campion, Peer review: crude and understudied, but indispensable, JAMA 272 (1994), pp. 96–9
The conclusion that was reached after a few annual scientific conferences published in the Journal of the American Medical Association as to the merit of peer-review were: “(i) blinding reviewers to authors’ identity does not usefully improve the quality of reviews, (ii) there is no association between reviewers signing their reviews and the quality of the review, (iii) passing reviewers’ comments to co-reviewers has no obvious effect on the quality of review, (iv) reviewers aged under 40—-write reviews of slightly better quality, (v) appreciable bias and parochialism exists in the review system. Finally, and perhaps most significantly, developing a useful instrument(s) to measure manuscript quality remains a huge challenge“. [and in the final analysis peer review] “can screen out [studies] that are poorly conceived, poorly designed, poorly executed, trivial, marginal, or uninterruptable.” No mean feat, really. But many scientists maintain that peer -review is a screen for quality and impact, not just for screening out bad science for funding agencies and for journals.
Neither Errol Friedberg, nor the authors of the congress proceedings seem to suggest alternatives. Rather, they present examinations of the process and its effect upon the final outcome. Friedberg also suggests that one constraint, that of page numbers in a journal, has been essentially removed with the advent of electronic publication, and thus more meritorious articles can now be published. Interestingly enough, many scientists — and journals — seem to value publication quotas, as those add prestige to those papers that do get accepted.
However, there are two things of which I’m certain: change, if any, will not come soon, but also we have not heard the last critique of the peer-review system.
Friedberg, E. (2010). Peer review of scientific papers—A never-ending conumdrum DNA Repair DOI: 10.1016/j.dnarep.2010.03.003
A promising meeting you may want to attend. Full disclosure: I’m in the program committee for this one.
ISMB SIG CALL FOR ABSTRACTS AND POSTERS – Deadline April 30
Metagenomics, Metadata and Meta-analysis / Biosharing
A Special Interest Group (SIG) organized by the Genomic Standards
Consortium (http://gensc.org)
at ISMB 2010 on July 9-10, 2010 in Boston, Massachusetts, USAURL: http://tinyurl.com/m3bio2010
There are now thousands of genomes and metagenomes available for study.
Interest in improved sampling of diverse environments (e.g. ocean, soil, sediment,
and a range of hosts) combined with advances in the development and application of
ultra-high throughput sequence methodologies are set to vastly accelerate the pace
at which new metagenomes are generated. The M3/Biosharing SIG will explore the latest
concepts, algorithms, tools, informatic pipelines, databases and standards that are being developed to cope with the analysis of vast quantities of metagenomic data. It will also seek to facilitate a broader dialogue among funders, journals, standards developers, technology developers and researchers on the critical issue of data
sharing within the metagenomics community and beyond through the
inaugural meeting of the BioSharing community (See http://biosharing.org
for a list of participating communities).Through two days of invited and contributed talks, panel discussions,
and flash talks associated with poster sessions, we aim to highlight
scientific advances in these fields and identify core computational
challenges facing the wider community. We invite you to submit extended
abstracts to be considered as talks or posters for the M3/Biosharing
SIG. The abstracts will be reviewed by the SIG program committee for
suitability and quality. Select abstracts will be published in a special
issue of the open access online journal Standards in Genomic Sciences
(http://standardsingenomics.org/).Topics include, but are not limited to:
M3
* Metagenome and microbiome studies of biological interest
* Metagenome annotation
* Consistent contextual (meta)data acquisition and storage in metagenomics
* Contextual (meta)data and sequence data correlation studies
* Computational infrastructures for metagenomic data processing
* Algorithms, data structures and database architecturesBioSharing
* Standard operating procedures, data standards. Ontologies and file formats
* Projects enabling open data sharing in science
* Data Sharing PoliciesConfirmed Plenary speakers include:
* Nikos Kyrpides, DOE Joint Genome Institute
* Michael Ashburner, University of Cambridge
* James Tiedje, Michigan State University
* Edward DeLong, Massachusetts Institute of Technology
* Ewan Birney, European Bioinformatics Institute
* Folker Meyer, Argonne National Laboratory
* Eric Alm, Massachusetts Institute of TechnologyCo-chairs:
* Edward DeLong, Massachusetts Institute of Technology
* Owen White, University of Maryland
* Dawn Field, NERC Centre for Ecology and Hydrology, UK
* Susanna-Assunta Sansone, European Bioinformatics InstituteImportant dates:
* April 30, 2010: talk and poster abstracts due
* May 7, 2010: notification of acceptance
* July 9-10, 2010: M3/Biosharing SIG eventFor more information including submission:
http://tinyurl.com/m3bio2010