There has been a lot of buzz recently about Wolfram’s new product, the Wolfram Alpha (WA). After attending a webinar on WA, I was given a preview account, and started messing around with it. In case you were wondering, that is the extent of my involvement with Wolfram Research, LLC, I don’t even have a Mathematica license. After playing around with WA for a few hours, I can safely say the following: it’s different, it’s incomplete, it’s idiosyncratic, and it’s funky cool. And no, it will not dethrone Google, nor does it aim to do so. Pish-posh.
It’s Different
Stephen Wolfram describes WA as a “computational knowledge engine”. It is not a web search engine: the information is curated and internal to WA, not searched over the web. Neither is WA an encyclopaedia: the information it provides on any topic is rudimentary, and mostly calculable. If you enter a country’s name, it will give you its GDP, GNP, size, population, but not its history. WA is more like an almanac with computational capabilities.
Yes, you can look up facts, like the GDP of Germany, the population of London, or the height of the Eiger. But the real power of WA lies in its ability to take data that can be numerically represented, and compute new relationships. A simple example is who has the larger population: Los-Angeles or New-York, and by how much?
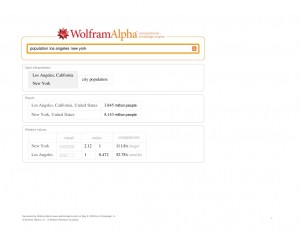
LA vs. NYC. Click to Enlarge
wolframalpha-populationlosangelesnewyork
It also shows comparative population growth over time.
The input screen is a single-line entry field, a-la Google, with quite a few links to examples, help docs, a blog, downloads (such as browser plugins) and an FAQ which is empty for now. The input field can supposedly take regular queries in English, and figure out a computable answer. You can also narrow down your query to a certain parameter, for example “males Canada Germany” will give you a comparison only of the male population of those two countries.
The output can be manipulated to show the results in different views. For example, you can show a weather forecast in Celsius or Fahrenheit. You can also show or hide results you may deem too detailed. You can also generate a PDF of the output, for download and archiving. However, the PDF is generated from the original query, and not from the modified output screen.
WA can be used to settle Guinness Book of World Records-like disputes, here is what “fastest car” brought me; you can also ask for the “highest mountain” or “oldest person”.
WA really kicks butt when it comes to math. But then again, it’s got Mathematica behind it, so in this context, it is basically a Mathematica front-end capable of deciphering natural language queries. No mean feat, really, but quite expected from the authors of Mathematica. Here is a thorough Mathematica-style analysis of the function sin(x)/cos(y). The PDF was rather large here, because of the graphics ( 13MB, another thing to fix, WA developers!), I whittled it down to manageable jpegs:

sin(x)*cos(x) p.1 Click to enlarge

sin(x)*cos(y) p 2. Click to enlarge

sin(x)*cos(y) p 3. Click to enlarge
And a matrix rotation:
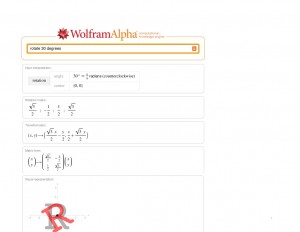
Matrix rotation, click to enlarge
Some everyday calculations, such as “compound interest,” or “body mass index” open a form where you can enter the parameters (principal, interest & maturity or height and weight, respectively) and receive a solution, including a graph over time for compound interest. When I entered “Moore’s Law”, I was pleasantly surprised to get form which lets me calculate the number of transistors per integrated circuits over time (Moore’s Law is a well-known empirical geeky law that states this number doubles every 18 months).
Anything to do with Biology? Yes. For starters, WA has a quite a few organisms. I typed “pea rose” and here is what I got:
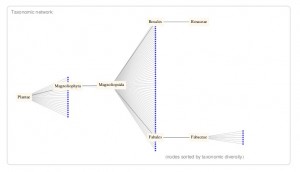
Pea vs. Rose. Click to enlarge
WA also hosts the sequence of the human genome (not sure if it’s Venter’s, Watson’s or NCBI’s). You can enter a DNA sequence and get exact matches, including genes. Seems like they are only hosting translatable genes: I could not find a tRNA I was looking for, and no intergenic regions. Also, it only gives exact matches, so not a lot of functionality there.
How about music? Yep. It even plays scales and chords; like my favorite, the D blues scale.
It’s incomplete
Some things seem to be missing, on a rather arbitrary basis. Here I need to explain something: it seems like every query word in WA is tagged as parts of a larger contextual semantic set. For example, “rose” can be a person’s name, but it can also be a plant. New York can be a city, or a state. WA makes intelligent guesses based on query context: if you query “population New York California” it will compare state populations, if you query “population New York San Diego” it will compare city populations.
That being said, yeast is tagged as a food, but not as an organism, so I could not compare yeast to pea on a phylogenetic tree like I did with rose and pea in the example above. By the way, rose can be a given name, or a surname, or a species, or a color, or a word, or a financial entity (stock ticker of Rosetta Resources Inc.). A rose in different semantic contexts in may not smell as sweet tho’ it be called by the same name.
Which reminds me, that “Romeo and Juliet” is tagged as a book or a movie (why not a play?). As a movie, WA brings us information on all three versions: George Cukor’s 1936 version, Franco Zephirelli’s 1968, and Baz Kuhrman’s 1996 (yeah, the crappy one, with DiCaprio). I could not get them side by side though, which is a shame: I like comparing movie versions, and who played whom.
In some cases, I couldn’t get graphs for functions where one of the variables was a denominator, and I am not sure why: if it’s because they are not handling zero division yet? Unlikely.
The main model organisms, Arabidopsis thaliana,(thale cress) Drosophila melanogaster (fruit fly), Escherichia coli (a bacterium) and Caenorhabditis elegans (nematode worm) are not listed. Strange, as although these are not exactly what the man on the street would think of as interesting species, they are central to life science, and I would expect them to be in a knowledge resource such as WA. Saccharomyces cerevisiae (baker’s yeast) and Danio rerio (zebra fish) are actually in there. If WA is interested in taking a slice out of the genomic browsing pie, it would do well to enter the genomes of those organisms as well. Then we could do some neat, if rudimentary, comparative genomics. As far as I could gather, only the human genome is in WA now. It would be cool to use WA for some quick & dirty comparative genomics.
One big point: I could not find how to drill deeper. Once I got, say, the schematic phylogenetic tree of two organisms, can I zoom in? Or once I get the GDPs of Germany and Canada side by side, can I look for private vs. public sector GDP? Trade deficit? I hope that once the documentation is in place, this would sort itself out.
It’s idiosyncratic
Small annoyances: imperial units and North American date formats are mostly the norm, except for scientific entries where SI standards are adhered to. Diseases are tagged as “cause of death”, so mortality statistics are given, but not morbidity.
Anyone who read A New Kind of Science knows about Wofram’s fascination with cellular automata. WA does cellular automata too: typing “rule 60” got me this:
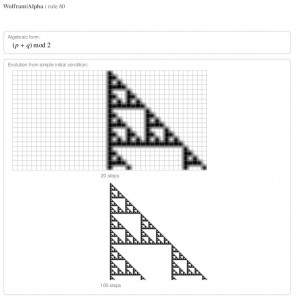
Rule 60 gives a cellular automaton. Click to enlarge.
So the phrase “rule (number)” automatically maps to Wofram’s idea of what a rule is, which is a cellular automaton rule.
It’s funky cool
Let me end by repeating where I started: WA is not a web search engine, and not an encyclopedia. No crowdsourcing or community-generated knowledge here. WA works mostly with curated sources, computable data, and hence allows for comparison. There is a bit of a learning curve regarding query syntax, and documentation is sorely lacking on that, but I hope it will be there by May 18, the announced release date. When all is said and done, I had lots of fun with WA. The single command line is very limiting, but Wolfram said in his webinar that they will provide some measure of an API to the public, and of course to whose who will purchase the commercial product. I hope that drill-down capabilities would be improved (or at least documented) because that is where the real strenght of this tool lies.
Oh, I almost forgot: you can compare apples and oranges. So there!
Update: for the slashdotters who asked about the “Meaning of Life”: not entirely unexpected:
Meaning of Life click to enlarge