Skin flick
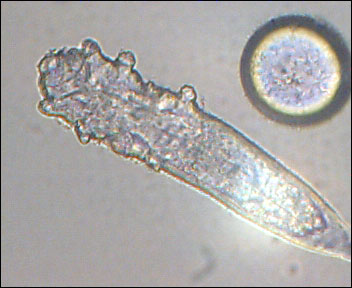
Interesting report in Science today about the human skin metagenome. The skin is a fairly large organ, and it is home to an estimated 1012 bacteria. It is the first barrier our body poses against pathogens, toxins, and sarcastic comments. An adult’s skin area is about 2m2, virtually all of it exposed to the outside world, picking up microbes, particles, small critters like the demodex (eyelash mite), and any residual matter from what we touch or touches us.
Title:
Calculate Your Body Surface Area
Description:
'Enter height, weight gender and age'
As an aside, the skin’s 2m2 surface area pales in comparison to that of the small intestine: 250m2, due to the huge number of villi used to absorb nutrients from the food; Also the lungs are estimated to have 160m2, with their sponge-like consistency.
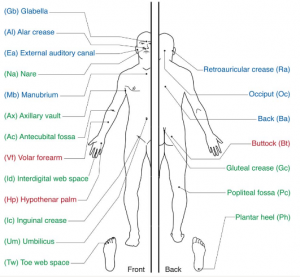
Until now, any study of the human skin microbiome was limited to those bacteria that could be cultured. The NIH has recently funded a $115M five year effort to study the human microbiome using metagenomic methods, that is, without the need to culture bacteria. As part of that, the initial species survey for skin is available now in Science. The researchers at the National Human Genome Research Institute in Bethesda, MD USA swabbed ten volunteers from different parts of their skin, and sequenced the 16S ribosmal RNA used for phylogenetic classification. They then looked at composition and the diversity of the bacterial communities in different areas of the skin. There are seven tie-ins for first place in diversity: behind the knee, on the heel, inner elbow, between the fingers, on the forearm, in the navel and the gluteal crease . The least diverse populations were on the back, and behind the ears(!?)
Another comparison they made was left-right symmetry, or rather the lack thereof. There are significant differences in bacterial population and diversity between the left and right sides. This was unexpected, and still unclear to why that is. Differences between “dry” “moist” and “oily” areas in the skin were also very pronounced, although that not entirely unexpected.
The goal of this report is to understand what is the normal bacterial population on the human skin, so we can understand what goes wrong in disease, an maybe be able to better diagnose skin disease. And not only skin diseases: the dreaded Methicillin resistant Staphylococcus aureus (MRSA) which causes thousands of hospital deaths each year lives benignly in our nasal cavity. Understanding its “ecological niche” can lead to a better way to combat it, or at least to keep it at bay.
One thing that was unclear to me was whether this project will go on to sample non prokaryal populations: i.e. fungi, viruses and microscopic mites. Some of the most bothersome skin infections, from ringworm to athlete’s foot are not caused by bacteria. Fungi, viruses and microscopic animals are also residents on out 2m2 of real estate.
Elizabeth A. Grice, Heidi H. Kong, Sean Conlan, Clayton B. Deming, Joie Davis, Alice C. Young, NISC Comparative Sequencing Program, Gerard G. Bouffard, Robert W. Blakesley, Patrick R. Murray, Eric D. Green, Maria L. Turner, & Julia A. Segre (2009). Topographical and Temporal Diversity of the Human Skin Microbiome Science, 324 (5931), 1190-1192 DOI: http://www.sciencemag.org/cgi/content/full/324/5931/1190
Comments are closed.