Quorum sensing
Social behavior is not exactly the first term that comes to mind with relation to microbes. After all, we assume a certain amount of intelligence and an ability to implement a behavioral pattern in response to peer actions. Humans, yes. Apes, yes. Birds of a feather flock together… so birds, yes. Ants and bees and other social insects, sure. But bacteria?
Yes, bacteria are social creatures: they can cooperate as a community. For example, many bacteria live in a biofilm, a tangled matrix of polymeric substances that includes proteins, DNA and polysaccharides. Biofilms constitute tough physical barriers that are immune to attacks by many antibiotics and other bacteriocidal agents. Indeed, many of the harder to treat infectious diseases are a result of the formation of biofilms in our bodies. A biofilm is analogous to a bunch of humans banding together, and deciding that instead of living in dispersed separate dwellings, they will all live together in a walled city that is easier to defend from attacks.
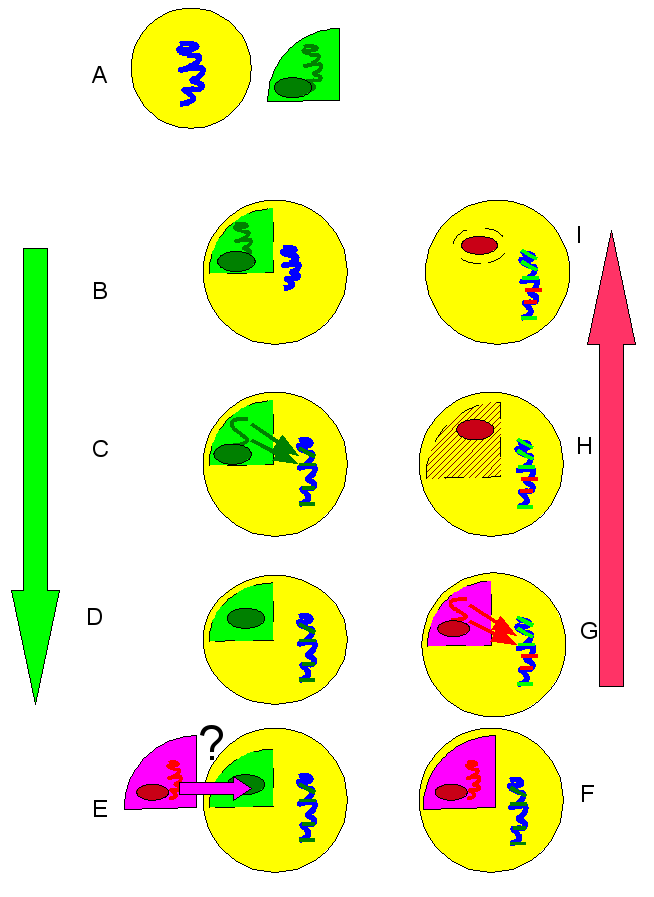
To achieve this cooperation, each bacterial cell starts by sending a signal. A molecule that says: “Yoohoo, I am here and I can help build a biofilm. Let me know if others are interested”. As more Bacteria send this molecular signal, its concentration in the environment grows. The sending bacterium also senses the environmental concentration of this signal. At some point, the concentration of the “yoohoo” signal reaches a certain threshold, and now each bacterium is convinced that rolling up its tiny sleeves and helping build a biofilm is actually a good use of its time and metabolic resources. The bacterial cell now begins to release biofilm building components, under the assumption that everybody around it is doing the same: after all, there is a lot of yoohoo signaling going on. This method of signalling is called quorum sensing (QS). Quorum sensing is used not only for biofilm construction, but for other group activities by bacteria. Secretion of virulence factors that damage the host, or molecules for scavenging nutrients. All these activities that are also community based.
Freeloaders
But wherever there is community work to be done, there is the danger of freeloaders: those who benefit from the labor of the community, but provide little or no input themselves. Are bacterial communities an exception? This question has been asked by several research groups, experimental and theoretical.
In 2007, Stephen Diggle and his colleagues have created two types of QS-related Pseudomonas aureginosa mutants. First, those who do not send the signal, hence they make no effort in propagating the information that a biofilm is being constructed (signal-negative). The second type produce the signal, but not the necessary products for constructing the biofilm (signal-blind). They then examined how well these mutants did alongside regular bacteria, in a stressful environment that facilitates the creation of biofilms. They started a culture with a small percentage os signal-negative and signal lind mutants (1-3% of the total population). Both types of cheating bacteria proliferated rather well, rising up to 45% and 66% of the populations respectively. But once cheats grew more common, their ability to proliferate of their fitness declines. Diggle and his colleagues attributed that to the decline in the number of cooperators that cannot support the cheats.
Why would cheating increase fitness, even of transiently? The answer is that producing both the quorum sensing signal, and the actual biofilm building components is metabolically costly. QS is therefore very sensitive to parasites: those strains that don’t have to produce signals nor the actual components will therefore benefit more than their hard working neighbors. Up to a point, that is.
The game of life. For life.
A recent theoretical study in PLoS-ONE examines the evolutionary fitness of hypothetical QS mutants that freeload. Note that this is theoretical: no Pseudomonas were harmed in this study.
Czaran and Hoekstra looked at the problem from an opposite point of view than that of Diggle. They asked whether QS individuals invade and proliferate in a non-QS population. To answer this question, they used a cellular automaton simulation. A cellular automaton is a grid in which the composing cells have different states (i.e. “full” or “empty”), and whose state depends on the neighboring cells’ state. Each time the grid is scanned, for each cell the neighboring cells determine that cell’s state in the next generation. Here is a simple cellular automaton called The Game of Life. In the Game of Life, each cell can be either “alive” or “dead”, depending on the number of neighboring live cells. A cellular automaton is therefore a good basis for simulating bacterial communities. In Czaran and Hoekstra’s simulation, each cell is a bacterium, that can be fully QS capable or QS incapable, or partially QS capable in different manners.
Ignoramuses, Liars and Voyeurs
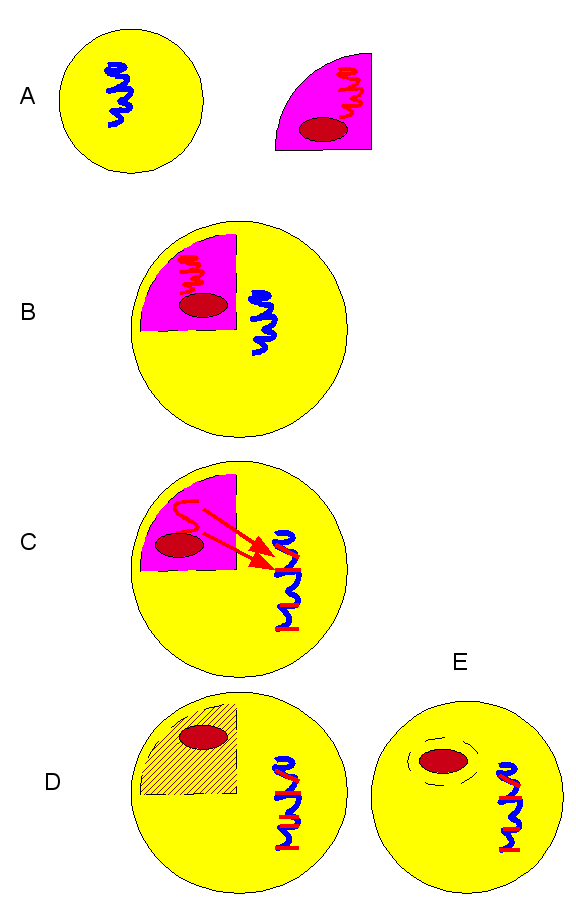
Creating strains in a computer is much easier than in real life, Czaran and Hoekstra used 3 loci for their simulated bacterial genomes. C for cooperation: production of a public good molecule, such as a polysaccharide for the biofilm. The other two for quorum sensing: locus S for producing the signal molecule (“yoohoo, I’m here”) and locus R for signal response, which includes the signal receptor and the signal transduction machinery that triggers the cooperative behaviour when the threshold signal concentration has been reached. The created 23 = 8 different strains based on the presence or absence of each active gene: Ignorant, Voyeur, Liar, Lame, Blunt, Shy, Vain, Honest. The Ignorant (csr) lives in complete solitude, and cannot participate in QS. The Honest (CSR) is a good QS citizen. The various others are freeloaders to some extent. For example, the Liar (cSr) produces the signal molecule, but not the actual response. Lame produces the quorum sensor and the response signal, but cannot produce the actual public-good (C) molecule.
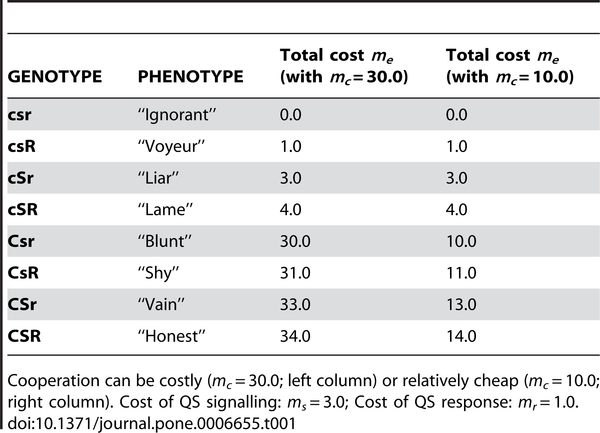
Table 1. The 8 possible genotypes of the cooperation-quorum sensing system and the corresponding total metabolic costs m(e) of gene expression.
They then ran the simulation using cellular automata. They started with mixtures of initial different populations, and ran the automata, with each cell’s response being a function of how it can respond (a liar cannot build a biofilm even though it asks everyone else to, while an Honest cannot help but sensing the signals and contributing). Since, as Diggle and colleagues have shown, being a good citizen is metabolically costly, Czaran and Hoekstra figured the metabolic cost in their simulations.
In a nutshell, Czaran and Hoekstra have shown that “both cooperation and the associated communication system can evolve, spread and persist in the population“. So being a good citizen pays off, and cooperation actually increases the fitness of the cooperative strains as opposed to the non-cooperative ones. This is a very elegant and informative simulation work, and I recommend reading it, since there is quite a bit more there than I have written about. My only complaint is that they did not provide some online resource to play around with seeding initial populations and seeing what happens to them after a multi-generational run. So just to psych you out a bit, here is a cellular automaton from YouTube:
And a biofilm, er, film:
Diggle, S., Griffin, A., Campbell, G., & West, S. (2007). Cooperation and conflict in quorum-sensing bacterial populations Nature, 450 (7168), 411-414 DOI: 10.1038/nature06279
Czárán T, & Hoekstra RF (2009). Microbial communication, cooperation and cheating: quorum sensing drives the evolution of cooperation in bacteria. PloS one, 4 (8) PMID: 19684853