Size matters. Life is Live.
1976: Prologue
In July 27, 1976, the American Legion, a US military veterans association, held a large meeting at the Bellevue Stratford Hotel a hotel in Philadelphia PA, celebrating the USA’s bicentennial year. Within 2 days, guests started falling ill with an atypical pneumonia. By the end of the week, 221 people were ill and 34 died in from an unidentified respiratory disease. Samples that were taken from the air conditioning’s cooling tower’s water yielded a formerly unknown bacterium. Legionella pneumophila was discovered, isolated and typified. “Legion Fever” or Legionnaire’s disease became a household name. It is estimated the US has between 10,000 and 50,000 cases of Legionelliosis each year. Legionella bacteria have been found to be typically in poorly maintained air-conditioning systems, whirlpool baths, indoor fountains, breathing apparatuses and any other warm water source where water is not constantly replaced or treated. The discovery of Legionella prompted new standards for maintaining such facilities and appliances.
1992-2002:”That’s no bacterium”
Fast forward to 1992, when several people become ill with pneumonia in the West Yorkshire town of Bradford. Because of the pattern of infection, the UK heath authorities suspected Legionellosis. Timothy Rowbotham from the English Public Health Laboratories had extensive experience with Legionella, and where to look for it. He collected samples from the water of a nearby cooling tower. No Legionella was found, but a small gram-positive cocccus-like microorganism was discovered infecting amoeba from the water in the cooling tower. Rowbotham named it Bradfordcoccus. Besides a name, very little was discovered about the new bug. The samples lay in the freezer for a few years, since Rowbotham could not culture it.
Ten years later, in 2002, work on Bradfordcoccus suddenly resumed when Richard Birtle brought samples from Rowbotham’s lab to the School of Medicine in Marseilles, to work with Didier Raoult on identifying pneumonia causing agents that grew in amoeba. Raoult’s lab specialized in intracellular bacteria and was experienced in isolating, culturing and identifying those elusive cells-within-cells. Samples from various different Legionella outbreaks were analyzed, and new strains of Legionella were identified. The Bradford sample was a frustration though: the typical way to identify bacteria is to isolate its DNA and then look for a specific marker genes, like a barcode, which identifies it. The gene customarily used is the 16S ribosomal RNA. This is the RNA that constitutes part of the ribosome, the complex protein-and-RNA machinery used by all living things to translate messenger RNA into proteins. Only Bradfordcoccus did not seem to have any 16SrRNA genes. All living creatures have rRNA genes, so after a frustrating year of vain attempts Raoult suspected that Bradfordcoccus’s cell wall was simply highly resistant to all the digestive agents they used to try and break it down and release the nucleic acids inside. It was time to take a much closer look at what was going on there. Raoult and Birtle prepared an electron microscope sample of amoebae infected with Bradfordcoccus. When the images came in they realized: that’s no bacterium they were looking at. Under a light microscope, the stained spots in the amoeba looked just like any other cocci type parasitic bacteria. But with the higher magnification and resolution of the electron microscope, the spots were shown to have a sharp hexagonal shape. This made it clear that they were looking not at a bacterium, but at a virus. Moreover, they were looking at the largest virus ever seen! At a whopping 0.75μm (micrometers), it was larger than quite a few bacteria.
That also explains the lack of 16rRNA: viruses do not have cellular translation machinery, including ribosomes and ribosomal RNA: they hijack that from their hosts. They named it the Amoeba polyphaga microbial mimicking virus, or mimivirus, or APMV.
2004. Genome: you gotta know what’s what
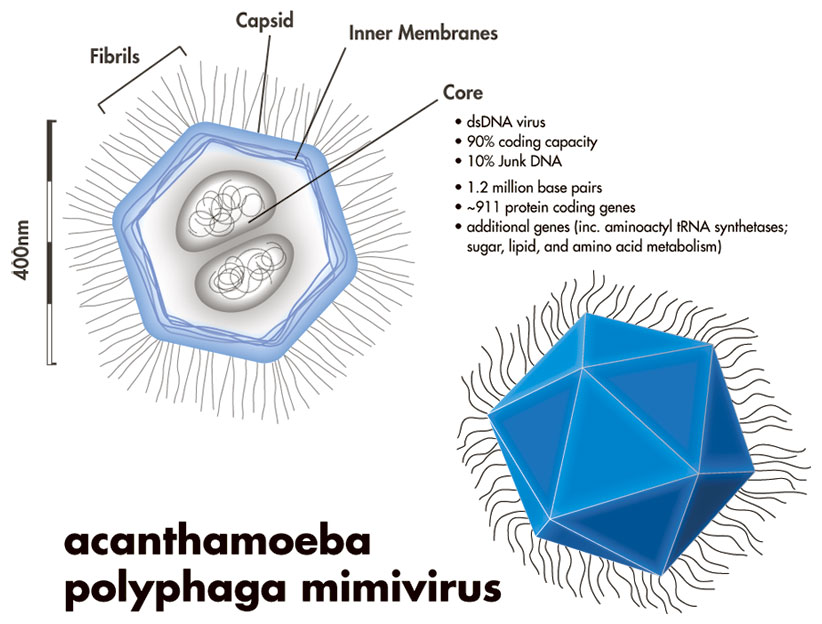
Year 2004. Jean-Michel Claverie’s and Didier Raoult’s groups publish the genome of Mimivirus in Science. Guess what: a big virus has a big genome. At 1,181,404 nucleotides, 911 protein coding genes and 6 strucutral RNA genes it is bigger than some 25 bacteria and archeae. The coding density — ratio of coding regions to non-coding regions is 90%, which is high even for viruses. This means that only 10% of the viral genome does not code for genes. So why is it so large? Apparently, there are lots of duplications, including one region which is 20% of the genomes length. About 35% of the virus’s genes have a homolog in the genome. This is higher than in most bacteria: it seems like the small genomic real-estate constraint — the need for compactness in the virus genomic DNA due to its small size — does not worry the mimivirus too much. Even with duplication, that leaves 400 original genes in the genome. That’s a lot of genes for a virus!
What’s in there? Lots of goodies, I cannot go over all of them here. For example, Mimi has quite a few enzymes that have to do with DNA repair and proofreading. That actually makes sense: if a virus has a small genome (HIV-2 has eight genes!), and a high replication error rate, many of the genomes would be duplicated from the template DNA strand, but only a few will be able to code for a whole virus. However, those few in ratio will still be quite a lot in number, so the virus can replicate itself at a rate that is higher than replacement, allowing it to proliferate. But the longer the genome, even with the same error frequency inserted in the replication process as in a small genome, the virus will accumulate a larger absolute number of errors per genome. The large absolute number of errors will rendering a higher ratio of newly transcribed genomes as non-operational. With a viral genome the size of a small bacterium, this means the virus may not be able to create viable copies of itself at the necessary replacement rate. Furthermore, the large number of duplicates may serve as a buffer against faulty viral DNA synthesis: if one copy of a gene is faulty, its viral homolog can still function.
2008-2009: The shape I’m in
A cutout of mimivirus is shown above. One theory holds that mimiviruses started as proto-nuclei during the emergence of eukaryotes, or nucleated cells. The viral DNA is organized in membrane-bound compartments, similar to a cell nucleus. At the same time, the fibrils wrapping the virus have peptidoglycan components, similar to bacteria.
Each vertex on the virus’s surface is the meeting point of 5 triangles. There is one special vertex on the surface of the mimivirus, called “stargate” or “starfish”. The starfish seems to play a role in injecting the viral DNA into the amoeba host.
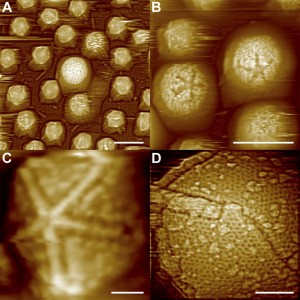
Atomic Force Microscope Images of Starfish-shaped Features on Defibered Mimiviruses. From: Structural Studies of the Giant Mimivirus Xiao C, Kuznetsov YG et al. (2009) PLoS Biology Vol. 7, No. 4, e92 doi:10.1371/journal.pbio.1000092
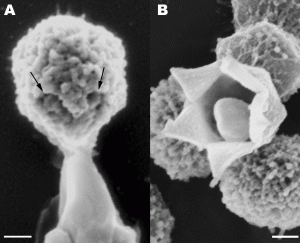
Stargate Opening From: Distinct DNA Exit and Packaging Portals in the Virus Acanthamoeba polyphaga mimivirus Zauberman N, Mutsafi Y et al. (2008) PLoS Biology Vol. 6, No. 5, e114 doi:10.1371/journal.pbio.0060114
2008: Sputnik comes to Mama
If all this was not enough, it seems that being a big virus also causes big problems: in 2008, Raoult and his colleagues discovered that a strain of mimivirus, called mamavirus, had a virus of its own! The small 50nm virus, (1000 smaller in dimaeter than mamavirus) was dubbed Sputnik, “little companion” in Russian. (Eugene Koonin is a co-author on the paper. Being originally from Russia, now at the NIH, I suspect he gave the new virus its name). Sputnik cannot infect the host amoeba by itself, but only when it is already infected with mimivirus particles. Sputnik seems to grow in mamavirus aggregates, called “virus factories” within the amoeba. It disturbs the growth and proliferation of the mamavirus. Sputnik has a genome of 18,343 bp, with only 21 protein coding genes, 3 of which cluster so closely with the mamavirus genes, it is almost certain they were derived from mama.
2009. Epilogue: what is life?
All these findings raise the question of the definition of Life. The common scientific definition includes an active metabolism, and common replicative and transcriptional machinery. Mimi does not seem to have a metabolism, nor does it have the full complement of replicative machinery, although it does have some tRNA genes, which are used to assemble proteins. It does have lots of genes, duplication, complex membranes, its own nucleus, and even its own parasite that not only acts as a pathogen of the virus, it also uses transfers from its host’s genome. Mimi may or may not be alive, but it certainly challenges our concepts of the living world, and causes us to think and redefine what we know and consider to be “alive”.
To finish up, here is the one hit wonder from the 80s, Opus. One of those songs that traumatized my youth, and were unfortunately burned into my psyche forever with the hot iron brand of incessant radio playings. It’s relevant though, just like Sputnik is relevant to Mamavirus, and Mamavirus is relevant to amoebae, and amobae are relevant to life. What is life? Life is Live.
La Scola, B., Desnues, C., Pagnier, I., Robert, C., Barrassi, L., Fournous, G., Merchat, M., Suzan-Monti, M., Forterre, P., Koonin, E., & Raoult, D. (2008). The virophage as a unique parasite of the giant mimivirus Nature, 455 (7209), 100-104 DOI: 10.1038/nature07218
Xiao, C., Kuznetsov, Y., Sun, S., Hafenstein, S., Kostyuchenko, V., Chipman, P., Suzan-Monti, M., Raoult, D., McPherson, A., & Rossmann, M. (2009). Structural Studies of the Giant Mimivirus PLoS Biology, 7 (4) DOI: 10.1371/journal.pbio.1000092
Zauberman, N., Mutsafi, Y., Halevy, D., Shimoni, E., Klein, E., Xiao, C., Sun, S., & Minsky, A. (2008). Distinct DNA Exit and Packaging Portals in the Virus Acanthamoeba polyphaga mimivirus PLoS Biology, 6 (5) DOI: 10.1371/journal.pbio.0060114
J.-M.Claverie, C. Abergel, H. Ogata (2009). Mimivirus Current Topics in Microbiology and Immunology, 328, 89-121 DOI: 10.1007/978-3-540-68618-7_3
Raoult, D. (2004). The 1.2-Megabase Genome Sequence of Mimivirus Science, 306 (5700), 1344-1350 DOI: 10.1126/science.1101485
Didier Raoult (2005). The Journey from Rikettsia to Mimivirus ASM News, 278-285 DOI: www.asm.org/ASM/files/ccLibraryFiles/FILENAME/000000001583/znw00605000278.pdf

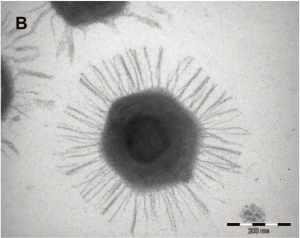
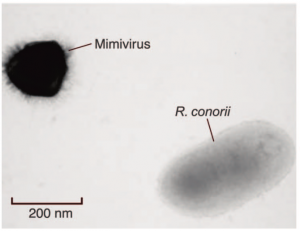


















Great writeup, thanks! All science should be presented as a story.