Of Mice and Men or: Revisiting the Ortholog Conjecture
I have posted quite a few times before about the acquisition of new functions by genes. In many cases a gene is duplicated, and one of the duplicates acquires a new function. This is one basic evolutionary mechanism of acquiring new functions.
Sometimes, gene duplication occurs within a species: part of the chromosome may be duplicated, causing one, a few, or many genes to have more copies of themselves within the species. The descendants of the duplicates and the original are homologous are they are descended from a common ancestor. This type of homology is called paralogy: a homology due to a duplication event (para == in parallel).
In another case, the genes can be homologous due to speciation: a new species (A1) diverges from the original (A0), carrying highly similar genetic loads. The gene for, say, brown eyes in A1 and the gene for brown eyes in A0 are also homologous: derived from the gene of hemoglobin in A0. This time, the homology is called orthology: it is not due to in-species duplication, but due to speciation itself (ortho == exact). The definitions of orthologs and paralogs were given by Walter Fitch in a seminal paper published in 1970.
One of the first protein structures to be solved was that of hemoglobin, the oxygen carrying protein complex in our blood. Scientists noticed that hemoglobin in jawed mammals has three different protein chains: alpha, beta and gamma. Their amino acid sequences were very similar, suggesting that the genes encoding for hemoglobin are highly similar, suggesting homology. Since all jawed mammals have hemoglobin, and they all had alpha, beta and gamma chains, the conclusion was that the duplication of the original genes happened in the common ancestor of jawed mammals, before they split up into different species. Hence, the alpha, beta and gamma chains in hemoglobin are paralogous: homologous due to duplication preceding speciation. However, gamma-hemoglobin was shown to have a different function than beta or alpha (more on that in a bit). The conclusion from this observation was the Ortholog Conjecture and it can be stated as follows: paralogs (reminder: homologs due to duplication) diverge in function more than orthologs (homologs due to speciation). A model was proposed for this observation: when genes duplicate within a species’ genome, there is less selective pressure on one copy to perform the same function. Thus, it can accumulate mutations and eventually adopt a different function. The ortholog conjecture states that paralogs mostly differ in function, whereas orthologs mostly do not. The ortholog conjecture is a very powerful statement because, if we have two proteins known to be orthologs, we can infer that they have the same function, whereas paralogs may not (if they had enough time to diverge). The ortholog conjecture is therefore a fundamental tenet in molecular phylogenetics, and is also a tool used to predict the function of proteins. If two homologous proteins are found out to be orthologs, then it is assumed they have the same (or highly similar) functionality.
A crack in the ortholog conjecture was formed in study published late 2009 in a paper published by Romain A. Studer and Marc Robinson-Rechavi. I blogged then about their study:
Romain A. Studer and Marc Robinson-Rechavi challenge common wisdom by publishing a study that says: “it ain’t necessarily so”. They look at three alternative models of molecular function evolution: (i) subfunctionalization after duplication; (ii) neofunctionalization after duplication; and (iii) the ‘alternative model’ of equal change after duplication or speciation. Subfunctionalization holds that after duplication, each of the two copies of the gene performs only a subset of the functions of the ancestral single copy. Neofunctionalization holds that one of the two genes possesses a new, selectively beneficial function that was absent in the population before the duplication. The ‘alternative model’ states that the gain of new function is not preferential to paralogs and that orthologs may gain new functions at the same rate that paralogs do.
Studer and Robinson-Rechavi claim that few studies have been made to study the scope of any of these proposed models. They then lay out study designs for doing so, challenging other evolutionary biologists (and themselves?) to conduct these studies and examine whether the common wisdom that orthologs maintain function while paralogs gain function. What I like about this paper is that it not only makes a strong case for challenging conventional wisdom, it also lays out a series of possible routes of study to be taken up by others.
Now two studies have widened this crack to a rather large crevasse. The first is a study by scientists in Indiana University. In a way, this new publication is a response to Studer & Robinson-Rechavi’s call to arms on points (i) and (ii). The IU scientists (the Radivojac lab and the Hahn lab at the School of Informatics at Indiana University, Bloomington, IN) examined hundreds of pairs of orthologous and paralogous genes from the mouse and human genomes. They then examined whether paralogs had a higher functional similarity, or rather orthologs. What they found certainly defied the ortholog conjecture:
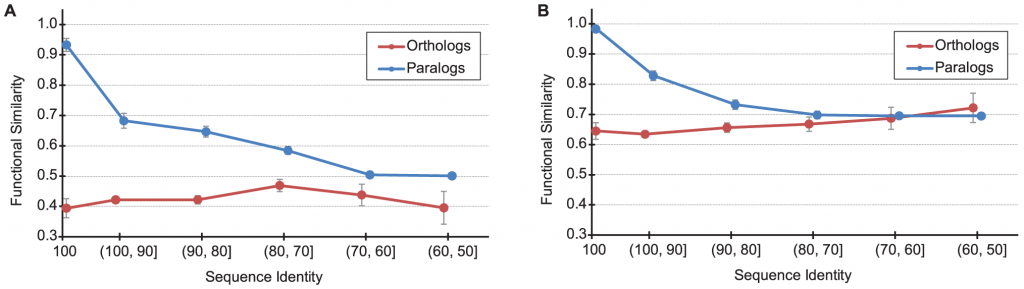
The relationship between functional similarity and sequence identity for human-mouse orthologs (red) and all paralogs (blue). (A) Biological pathway (B) molecular function. From PLoS Comput Biol 7(6): e1002073 under CC licence.
But before we explain the results, a word about function. The function of a protein has several aspects which are context-dependent; two important ones are the molecular function of the protein, and the biological process in which it participates. For example, the molecular function of all hemoglobins is noted as oxygen binding and oxygen transport. However, they are different in the processes, or pathways, in which they participate: gamma-hemoglobin participates in the transport of oxygen in the fetus. The complex which contains gamma-hemoglobin has a higher affinity to oxygen, and thus able to extract oxygen in the placenta from the maternal oxygenated hemoglobin and transport it to the fetus.
Now we can explain the figure above. Graph (A) above shows the functional similarity for the biological pathway aspect and how it is affected by the sequence identities of the hundreds of orthologs (red) and paralogs (blue) examined between human and mouse. Graph (B) shows the functional similarity of the molecular function aspect.
The X-axis is the sequence identity percentage between any pair of sequences: the higher the percent identity, the less divergent are the sequences, the more inclined we should be to think that the pair of proteins performs the same function since they diverged less. The Y-axis shows the fraction of functional similarity. Looking at graph (B) above, we see that paralogs which are 100% identical, have (almost always) the same function . But sequences of orthlogous proteins between human and mouse have only about 65% functional similarity, on average. What does that mean? In the database they looked at, each gene has a set of words associated with it, describing what it does. The IU scientists found that only about 65% of the keywords in orthologous sequence pairs overlapped, on average. Whereas for paralogs 100% overlapped. And those are for sequences which are identical! This means that even if we find identical protein sequences in human and in mouse, it does not mean that they have the same molecular function. On the other hand, paralogs, will generally have more similar functions. So the ortholog conjecture has been stood on its head here: paralogs are the ones that would generally have the same function, whereas orthologs diverge more in function. This holds true for up to about 50% sequence identity, when the picture seems to reverse itself. Graph (A) depicts the differences in the biological pathway aspect. Here, the differences are even more striking. The paralogs which are 90-100% identical between human and mouse participate in almost exactly the same pathways in both organisms. But orthologous proteins which are 90-100% identical the functional similarity is much lower: only about 65%.
![]()
So what does this all mean?
First, it means that, at least between human and mouse, paralogs are better predictors of function than orthologs. And why would that be? To answer this question, let’s look closer at the graphs above. Note that while for paralogs the functional similarity decreases rapidly with sequence similarity, for orthologs the functional similarity remains roughly the same no matter how similar or different the orthologs are to each other, and even when they are 100% identical their functions vary to some extent! The reason: the experimental study of function in two human and in mouse takes place in different contexts. The species-specific context is what causes the differences in annotation, and in the overall function. Also, all the orthologs in the study are of the same age, dating back to the human-mouse lineage split 75 million years ago. The paralogs predate that split, and may be of different ages: the split may predate the human / mouse split by 10 million years, 100 million years, or 1 billion years. Thus orthologs, regardless of their actual sequence similarity, have the same age, and paralogs do not. But why should proteins of the same age share the same level of (not so high) functional similarity? The authors of the study reply:
While there is no direct role for “time” in evolution that is not tied to mutation, we suggest that what time represents here is the evolution of the cellular context: the sum of the evolutionary changes over all of the directly and indirectly interacting molecules. If this context evolves at a steady rate (i.e. the average amount of functional change among all of the interacting molecules remains relatively constant), then protein function will appear to evolve at a steady rate, a rate largely disconnected from the level of an individual protein’s sequence divergence. — PLoS Comput Biol, Vol. 7, No. 6.
The strongest evidence they find for this hypothesis, is that even proteins with 100% are annotated differently. To wit:
For example, Liao and Zhang [50] found that >20% of genes that are essential for viability in humans are not essential in mouse. It is unlikely that changes to the proteins themselves have made them essential or not, but rather that their context in cellular and organismal networks has evolved. —ibid.
The proteins may not have changed substantially, but their environment changed, giving them a different role. Think about changing jobs after moving to a new place where there is no employer providing your exact old job you were used to. You may have been an embedded systems programmer, but now you are a website programmer. So context goes a long way to explain changes in ortholog function.
Interestingly, about a month after the IU paper was published, another paper from the Robinson-Rechavi lab was published, which also talks about homologs between human and mouse. In this study Gharib and Robinson-Rechavi reviewed previous literature listing several types of functional divergence of orthologs between human and mouse. They had some additional findings. For example, about 11% of the orthologous genes were alternatively spliced, meaning that the end products, proteins, were different between human and mouse. They also listed specific phenotypic effects: genes which are linked to diseases in humans, but mutations in their mouse orthologs have no effects on mice. They cite studies that found that over 20% of genes which are essential in human are non-essential in mice (an essential gene is just that: if the organism does not have it, or it is mutated, the effects are fatal, and the organism does not develop past very early stages). Their literature review concluded that 10-20% of ortholog pairs between human and mouse cannot be used for functional transfer. The IU study implies a higher percentage. Both studies conclude that a common practice in molecular evolution studies, the use of orthologs to infer function, should be seriously looked at.
(Full disclosure: Dr. Radivojac & I are collaborators, although our collaboration is unrelated to this study).
Nehrt, N., Clark, W., Radivojac, P., & Hahn, M. (2011). Testing the Ortholog Conjecture with Comparative Functional Genomic Data from Mammals PLoS Computational Biology, 7 (6) DOI: 10.1371/journal.pcbi.1002073
Gharib, W., & Robinson-Rechavi, M. (2011). When orthologs diverge between human and mouse Briefings in Bioinformatics DOI: 10.1093/bib/bbr031
Fitch, W. (1970). Distinguishing Homologous from Analogous Proteins Systematic Zoology, 19 (2) DOI: 10.2307/2412448
Comments are closed.


















