I am fascinated with zombies. Always have been, but even more so since I took an interest in microbiology. The zombie apocalypse is the best known and best chronicled viral infection which hasn’t happened. But it could happen any day, so stock up on non-perishable food, medical supplies, water purification tablets, chainsaws, machetes, baseball bats, crossbows, semi-automatic firearms, and as many disposable acquaintances as you can get hold of. Signs of the zombie apocalypse may include your significant other turning a putrid-greenish shade of rotten and trying to chew your neck off. Beware of the differential diagnosis that they might just be using a bit too much makeup, are not wearing deodorant and feeling flirtatious. Remember to eliminate that possibility before cracking your beloved’s head open with a crowbar.

Here are three interesting studies all having to do with zombies.
The Zombie Roach
One problem that has to do with zombification is the loss free will. Do zombies have free will? More to the point, do humans have free will? 28 Days Later and both versions of Dawn of the Dead have survivors finding refuge from the zombie apocalypse at shopping centers. While at the shopping center, the survivors copiously consume the goods in the stores, making you think who really is the mindlessly-obsessed drone lacking free will. In two papers entitled A Wasp Manipulates Neuronal Activity in the Sub-Esophageal Ganglion to Decrease the Drive for Walking in Its Cockroach Prey and On predatory wasps and zombie cockroaches — Investigations of “free will” and spontaneous behavior in insects, Ram Gal and Frederic Libersat from Ben Gurion University explore free will in cockroaches. Do cockroaches have free will, or are they just sophisticated automatons? And where do we draw the line between the two? Gal and Libersat use the following definition for free will: the expression of patterns of “endogenously-generated spontaneous behavior”. That is, a behavior which has a pattern (i.e. not just random fluctuations) and must come from within (i.e. not entirely in response to external stimuli). They cite studies where such behavior — which they define as a “precursor of free will in insects” — is observed. They then show how this behavior is removed from cockroaches when the roaches are attacked by a wasp. Their description of the process is so colorful, I shall simply reproduce it here. This is some of the most delightful and engaging prose I have ever read in a scientific paper.
Unlike most other parasitoids, this tropical Ampulicine wasp does not simply paralyze its prey to immobilize it. Instead, it stings a cockroach in the head (Fig. 1A) and injects a neurotoxic venom cocktail directly inside the cerebral ganglia (Fig. 1B). This turns the cockroach, metaphorically, into a submissive ‘zombie’: it gradually enters a long-lasting hypokinetic state, during which it becomes unresponsive to aversive stimuli and fails to self-initiate walking or escape behaviors. Although the stung cockroach is not paralyzed, it allows the wasp to cut both its antennae and drink hemolymph from the cut ends. The wasp then grabs one of the antennal stumps and pulls backwards, leading its prey into a pre-selected nest. The intoxicated cockroach, rather than fighting or fleeing its predator, actually follows the wasp submissively. In doing so it demonstrates a completely normal walking pattern, as if it was a dog led by his Master’s leash. The wasp then lays one egg on the cockroach’s leg, seals the nest and leaves the lethargic prey inside, still alive but powerless to escape under the influence of the venom. As the wasp larva hatches from the egg, it penetrates through the cockroach’s cuticle and feeds on its internal organs for several more days. Only then, roughly five days after the sting, does the cockroach finally die and the larva pupates inside its abdomen, safe from predators outside the nest.
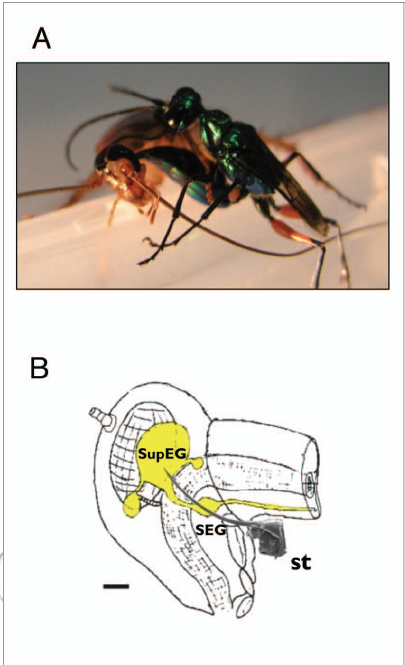
A. The parasitoid Jewel Wasp A. compressa stings its cockroach prey inside the head. B. Schematic drawing of the cerebral nervous system (yellow) inside the cockroach's head capsule. the wasp's stinger (st., scanning electron micrograph drawn to scale) reaches to inject venom into both cerebral ganglia, namely the supra-esophageal ganglion (SupEG) and sub-esophageal ganglion (SEG). Scale bar: 0.5 mm. Source: Commun Integr Biol. 2010 Sep-Oct; 3(5): 458–461.
What they also found was that the venom injection does not affect the roach’s muscle, motor neurons or sensory neurons. Those are all intact. Stung cockroaches will walk slower and submissively follow their wasp mistress, but if placed in water they will start paddling frantically like unzombified cockroaches trying to save themselves from drowning. Indeed, the zombies paddle as frantically as non-zombies, but for a much shorter time, as if they despair quicker.
But ah, you say — these cockroaches are not “real” zombies are they? Or rather, not the type of zombies from which the zombie apocalypse would be created. After all, a zombified roach is not infectious, and thus the disease would not spread from roach-to-roach in exponentially rising numbers by having mad roaches bite each other. In fact, the roach becomes catatonic and subservient to its mistress, more like the original Voodoo zombie, a reanimated corpse under the control of a shaman.
So how about molecular zombies?
Consider plants. Not zombie plants, but zombie genes in plants. Actually, transposable elements or TEs. TEs are DNA elements that self-replicate, in their own genomes or may be transferred into other genomes. Most eukaryotes have them. Indeed, 17% of the human genome is composed of Long Interspersed Integrated Elements or LINES. LINES simply replicate through the genome, and their contribution, if any, to the fitness of the organism is not known. some TEs are like internal viruses: replicating, adding their sequence, but little else, to their “host” genome. Only by increasing their numbers, they may at some point change their host genome. They may write themselves into some vital piece of DNA, and cause genetic damage that, if it does not kill the carrier while still an embryo, may cause long-term debilitating damage to the species by fixing itself in the population and moving down the generations. The genetic load of TEs in plants is huge: in Maize they make up the majority of the genome, with 85% of the genome coding for TE genes.

But the self-selection of TEs to spread in the genome is also their undoing. Small pieces of RNA derived from TEs are used by the host to target full TEs and degrade them through a group of processes known collectively as RNA silencing. Some TEs do generate an RNA or a protein end product. Those smaller, derived aberrant TEs were nicknamed ‘zombies’ in a paper by Damon Lish from University of California, Berkeley and Jeffrey Bennetzen from the University of Georgia, Athens. There is an ongoing battle between the TEs and the zombie TEs, with the latter silencing the former, yet metaphorically feeding off the original TEs existence. If the ‘live’ TEs did not exist, the ‘zombie’ version of them would not either. The more copies of TEs a genome has, the more zombie TEs it will have. TEs inserting themselves near active genes may subject these genes to silencing by zombie TEs. This ability of plants to silence their own genes by shuffling TEs around generates new patterns of gene expression. Also, the same genes that may be inhibited due to their proximity to TEs may, at some point, re-express themselves if the TE is removed from that position in the chromosomse. So zombie TEs, which are a form of defense against viral TEs that cause chromosome damage, are also a mechanism for generating new genes and expression patterns: an evolutionary tool.

Transposable elements ("jumping genes") are responsible for the different colors in Indian corn. TEs are controlled by their "zombie offspring": short interfereing RNAs whose sequences comlement those of the TEs, silencing TE expression.
Finally, there are the zombie ants
I have written about zombie ants before:
Here is a parasitic fungus that infects ants. The infected ant wanders away from its nest; the ant then reaches a leaf or another plant part. The fungus makes the ant to bite the leaf so powerfully, it hangs from the leaf until it eventually dies; and then (excited-by-gross-stuff six-year old emerging): the fungus grows an upside down stalk out of the dead ant’s head, releasing spores that fall to the ground. The spores are then picked up by ants that walk over them, causing them to wander away from the nest, bite other leaves… Ad nauseam. Wow.
Briefly, once infected, the ant’s behavior is hijacked to act as a delivery system for the fungus, which is finding a good location to die and infect more ants.
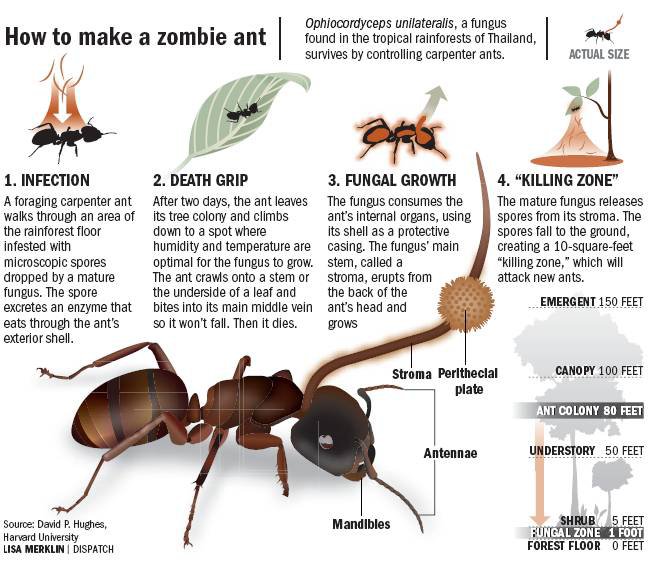
Ophiocordyceps unilateralis is the fungus that wreaks this havoc on the ants. Indeed, an O. unilateralis infection in ants is the closest parallel we can find to the human zombie apocalypse. The infection changes the ant’s behavior to infect more ants, and can decimate whole colonies.
But is there only one species of fungus? Harry Evans, Simon Elliot and David Hughes were inrigued by the original description of Torrubia unilateralis, as it was called at the time. In 1865, Louis René Tulasne a French mycologist, described a leaf-cutting and as the host for the fungus. His brother, Charles Tulasne, worked with him and illustrated their findings. Charles’s drawing of an infected ant does not depict the leaf-cutter: rather, it appears to be a carpenter ant, with its characteristic spines. The fungus has only ever been found infecting carpenter ants. Could Louis have made a mistake? Maybe there other species of ant-zombifying fungi out there?
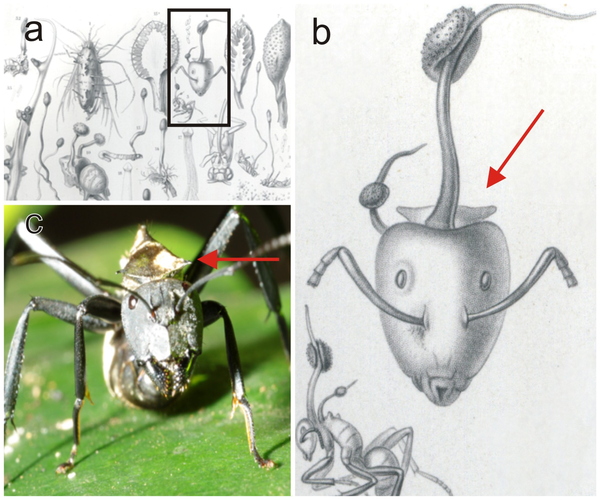
a. Original plate from the 1865 Selecta Fungorum Carpologia of the Tulasne brothers, illustrating the holotype of Ophiocordyceps (Torrubia) unilateralis and said to be on the leaf-cutting ant, Atta cephalotes; b. Detail from plate showing the distinctive pronotal plate of Camponotus sericeiventris, as well as a side view of the host which is clearly a carpenter ant and not a leaf-cutter; compare with c. Live worker of C. sericeiventris showing the spines on the pronotal plate (arrow). Source: doi:10.1371/journal.pone.0017024.g001
Indeed there are, and the authors of this study found four new species in the Brazilian rain forest, all distinct by shape and development. All infecting carpenter ants, (no leaf cutter ants found to be infected yet!) but four different species of carpenter ants: Camponotus rufipes, C. balzani, C. melanoticus and C. novogranadensis — are each attacked by a distinct species of Ophiocordyceps. Plenty of great pictures in the paper describing the differences between the ant-zombifying fungi.
Happy zombie-bashing! (Warning: Pop-up link to an external flash game).
Gal, R., & Libersat, F. (2010). A Wasp Manipulates Neuronal Activity in the Sub-Esophageal Ganglion to Decrease the Drive for Walking in Its Cockroach Prey PLoS ONE, 5 (4) DOI: 10.1371/journal.pone.0010019
Gal, R., & Libersat, F. (2010). On predatory wasps and zombie cockroaches: Investigations of free will and spontaneous behavior in insects Communicative & Integrative Biology, 3 (5), 458-461 DOI: 10.4161/cib.3.5.12472
Lisch, D., & Bennetzen, J. (2011). Transposable element origins of epigenetic gene regulation Current Opinion in Plant Biology, 14 (2), 156-161 DOI: 10.1016/j.pbi.2011.01.003
Evans, H., Elliot, S., & Hughes, D. (2011). Hidden Diversity Behind the Zombie-Ant Fungus Ophiocordyceps unilateralis: Four New Species Described from Carpenter Ants in Minas Gerais, Brazil PLoS ONE, 6 (3) DOI: 10.1371/journal.pone.0017024


















 .
.