A Flurry of Red and Green
UPDATE: I submitted this post to the National Evolutionary Synthesis Center’s sponsored contest for a travel award to ScienceOnline2010. Let’s see how it goes… #scio10
In a previous post about Hatena we saw what might very well be the beginning of a (beautiful?) [:ttip=”symbiosis where one partner lives inside the cell of the other” id=”10″]endosymbiotic[:/ttip] relationship: a unicellular predator swallows a microalga, resulting in physiological changes to both, and the resulting endosymbiont is now a [:ttip=”Uses light to synthesize food E.g. plants, algae” id=”phototroph”]phototroph[:/ttip], rather than a predator. “endo” – inside “symbiosis” – life together. Endosymbionts live out their symbiosis inside the host’s cells.
In this post I would like to fast-forward to another part of the endosymbiotic movie. We will see how endosymbiosis contributes to evolution much more than we thought. But first, some background information.
Primary and secondary endosymbiosis
Primary endosymbiosis happens when one free living organism engulfs another, resulting in a [:ttip=” Mutualism is a biological interaction between organisms, where each individual derives a benefit” id=”mutualism”]mutualistic[:/ttip] relationship. Secondary endosymbiosis is the process of engulfing another free-living organism that already went through primary endosymbiosis. Such is the case of Hatena: the algal endosymbiont provides the photosynthetic capability and light sensitivity (acquired by primary endosymbiosis), while the host provides motility and a cozy stable home: its cell. [:ttip=”Plant and algal organelles that manufacture and storage of important chemical compounds” id=”plast”]Plastids[:/ttip] are organelles that harvest light, manufacture pigments, store food and perform various other functions in plants and algae. Plastids are thought to be photosynthetic microbes that were acquired by primary and then secondary endosymbiosis: they have chromosomes encoding their own DNA transcription and translation machinery, as well as some other genes. One strong evidence for secondary rather than primary endosymbiosis is the number of membranes surrounding plastids: 3 membrane layers in algae, 2 in plants, strongly suggesting successive endosymbiotic events. Another evidence is molecular: most of the proteins needed for plastids to function are encoded in the host’s nucleus. How and why did the genes travel from the endosymbionts to the host?
Nobody is really sure yet, but here is a working hypothesis: once endosymbiosis occurs, the genome of the endosymbiont becomes mostly redundant. After all, the host takes care of most of the endosymbiont’s nutritional and metabolic needs, and maintains a stable environment in the cell. About 30% of a typical microbial genome is dedicated to genes that stabilize its internal environment in response to events in the external one. Most or all of these genes become redundant once the microbe in question becomes an endosymbiont, and enjoys the hospitality of its host, trusting it to maintain a controlled environment. They either disappear or migrate to the nucleus of the host.
Diatoms: hosting more types of algae for longer that you think
Diatoms are microscopic algae, so named because they are often shaped from two symmetric lobes — hence “diatoms”. They are photosynthetic, and are thought to compose most of the [:ttip=”‘plant’, photosynthetic plankton” id=”phytoplankton”]phytoplankton[:/ttip] biomass.
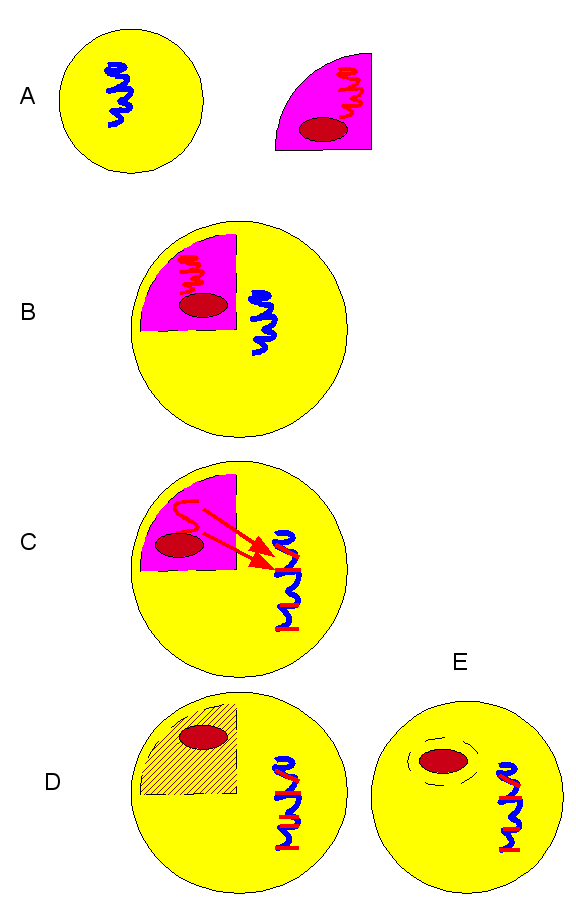
It has been known for a while that diatoms acquired their plastids by a process of secondary endosymbiosis with red algae. The commonly accepted sequence of events for the diatom / red algae endosymbiotic time-line is shown here:
(A) historical diatom (yellow) and red-algae: red ellipse is a generic plastid; (B) algal endosymbiont in diatom; (C) gene migration from alga to the diatom’s nucleal DNA; (D-E) algal cell mostly gone, only the plastid remains.
This is what Ahmed Moustafa and his colleagues also thought about the acquisition of chloroplasts by diatoms. They therefore set out to look for genes of red algae in the nuclear DNA of two diatom species whose genomes have been sequenced. To their surprise they discovered that 70% of the algal origin genes in the diatom were from green algae lineages, not red algae. However, there are no green algae-originating plastids in those diatoms. In particular, there were some genes that exist in the chloroplasts of red algae, but not in the secondary endosymbiotic chloroplasts in diatoms. So what happened? Why is the host’s genome “mostly green” instead of “all red”?
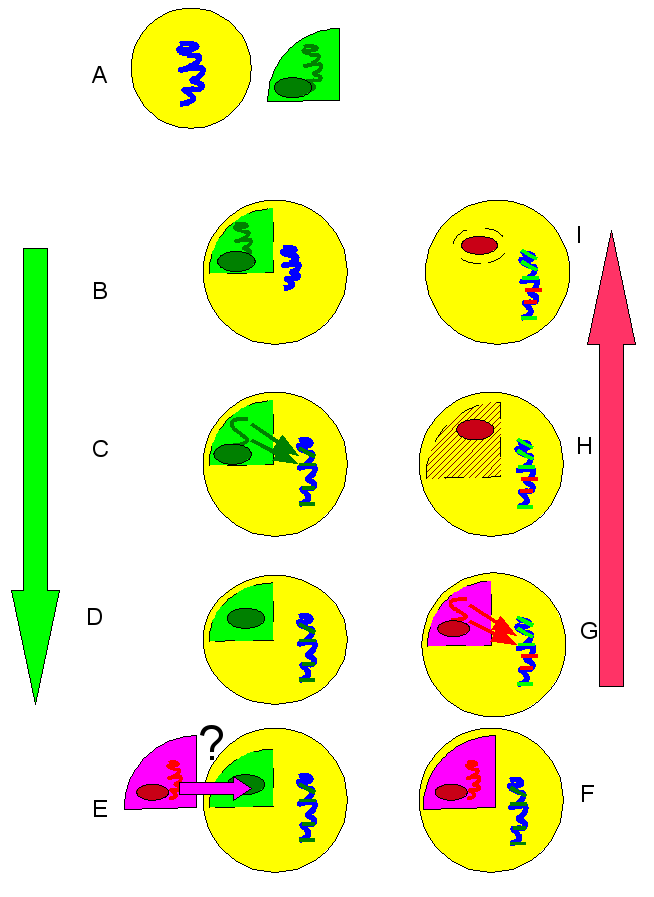
The answer that Moustafa and colleagues proposed was that these diatoms used to have plastids of green algae lineage. The genes that migrated to the diatom nuclear DNA are therefore green in origin. Over evolutionary time, for reasons unknown, red algae endosymbionts displaced the green ones. Many of the red genes that would have otherwise migrated to the nucleus already had their places take by green genes, and were simply lost.
A-D: first sequence of events: endosymbiosis of green algae, including gene migration to diatom nucleus; (E) displacement of green algae by red, through some unknown mechanism; (F-I): endosymbiosis of red algae, including gene migration to nucleus. Nucleus now has a mixture of green lineage and red lineage genes.
Many questions remain open: why did this replacement take place? How prevalent is it? The researchers only looked at two diatom species, whose genomes have been sequenced. One way to answer this question would be a [:ttip=”the study of genetic material recovered directly from environmental samples” id=”metag”]metagenomic[:/ttip] analysis of a diatom population. This would mean analyzing samples of DNA sequences taken from many different diatom species, to get a picture of the frequency of red versus green endosymbiont lineage genes in many more diatom genomes. Also, why would one set of endosymbionts be displaced by another? What is the evolutionary time-line in which the endosymbiosis / displacement process occurs? What, if anything, triggers this replacement?
This finding sheds light upon a larger question in evolutionary biology: how big is the role of endosymbiosis in evolution? How many of an organism’s genes are acquired from other organisms? It seems that with this study, the importance of endosymbiosis as a contributor of genes, just went up a notch: we see yet a few more cross-growths between the not-so-separate branches of the tree of life.
Finally, A flurry of Red and Green by The Dreamer and the Sleeper covered by Karys Rhea. The webcam self-shoot is grainy, and the sound is not much better than a laptop microphone. But Karys Rhea’s singing shines through.
Moustafa, A., Beszteri, B., Maier, U., Bowler, C., Valentin, K., & Bhattacharya, D. (2009). Genomic Footprints of a Cryptic Plastid Endosymbiosis in Diatoms Science, 324 (5935), 1724-1726 DOI: 10.1126/science.1172983






















Man my life sucks: I just typed a detailed post with citations about why it’d be weird if diatoms really did have green plastids before red, and suggesting perhaps the green was a temporary tertiary endosymbiont from which some genes were transferred followed by its disappearance… and then I missed the number verification thing and lost my comment forever. Now I’m really pissed. Deep breath…
But since I actually bothered to dig out the relevant citations, in pt form:
– chromalveolate hypothesis – common red algal plastid origin for stramenopiles (incl diatoms), alveolates and ‘crhaptophytes’. Even if not true, still most parsimonious to assume common origin for ochrophyte plastid, especially since some of the non-photosynthetic basal stramenopiles have evidence of plastids in their past. This common endosymbiosis. was red; green makes no sense. Separate endosymbiosis for diatom original plastid would be unparsimonious…
(for good review + diagram of plastid endosymbioses in euk history: Keeling 2004 Am J Bot)
– karlodinium (dinoflagellate) had tertiary endosymb with haptophyte; thereby having hapto plastid. It lost original plastid, but interestingly the new plastid-targetting genes are chimeric, mixed dino and hapto origin. (Patron, Waller & Keeling 2006 J Mol Biol)
-> perhaps in diatoms something similar happened, but in reverse: original plastid red algal, then temporary green algal tert. endosymbiont w partial gene transfer, then loss of green plastid?
Could be BS though, as I’m not a molecular biologist… >_>
Glad to see someone else write stuff about protists!
Cheers,
-Psi-
(hopefully this thing won’t eat my comment this time…not at this insane hour of the night anyway… grr!)
@Psi Wavefunction
Might be a good idea to draft long comments locally, then cut & paste to comment posting site. Sorry about your comments.
Thos are some very interesting alternative explanations. I am really quite new to endosymbiosis, so I cannot provide a good critique of yours vs. Moustafa’s suggestions. To be fair, I don’t think Moustafa, nor I ever suggested these were separate events: just that the green one may have occurred earlier, as you suggested with tertiary endosymbiosis. (Well, first there was red, then temporary green, then red only again: so temporary green did occur earlier than *current* red).