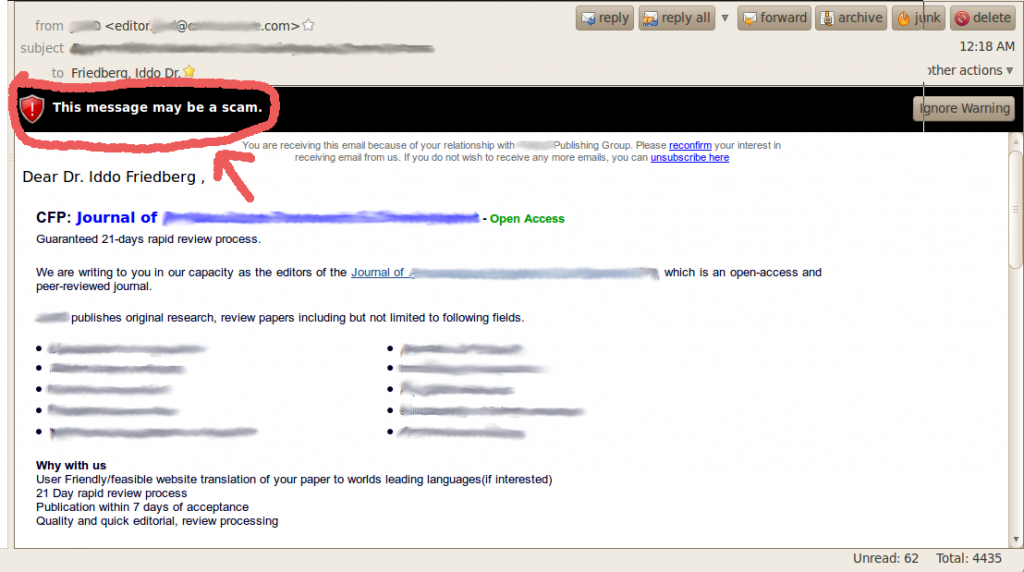
This came in my inbox today: solicitation to submit manuscripts to a journal completely outside my area of interest and proficiency. My email client software had an interesting reaction to it:
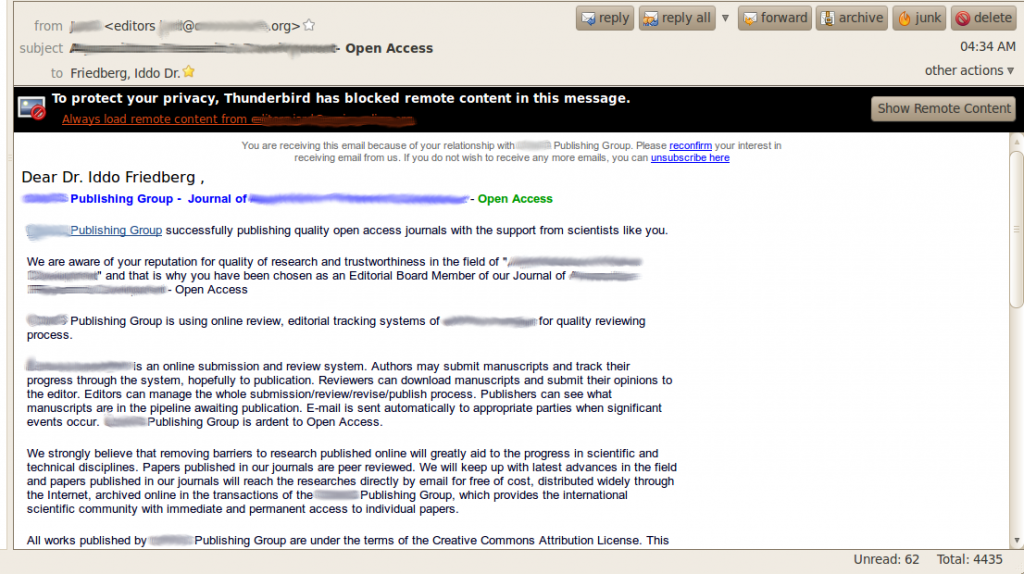
The next email in my inbox was this:
OK, that’s taking things a bit too far. Random solicitation of editors (did I mention that was not my field?) does not lend credibility to the publisher.
Which brings us to a nice post by Kevin Karplus about the not-so-savory side of open access publishing, or the “author-pays” model. First, a clarification of terms: Open Access means access to research publications by the general public without needing to pay for access through a personal or institutional subscription. Author-pays would mean that publication fees are covered by the author. The two are not necessarily cojoined. There are plenty of closed-access publications whose page and figure charges run as high or even higher than Open Access publications. But in the case of Open Access, the revenue does not come from subscriptions. The only other sources are advertisements and author fees. Since many Open access publications are web-only and are quite specialized, advertisements are not a practical source of revenue. This leaves us with author fees. So to publish open access you pretty much need to have money. If you happen to fall on hard times and you lose some funding, or need to skimp for a while, author-pays publishing is becomes less of an option, despite supposed hardship fee waivers. Since more journals are adopting the Open-Access model, many publications venues are being closed off to poorly funded researchers.
But I would like to address a point that was minor in Kevin’s post, yet is quite important: the wrongly perceived association of Open-Access with the cheapening of scientific publication. And by “cheapening” I do not mean the lowering of costs, although publication costs for web-only publications are considerably lower than those for print. I meant cheapening in the figurative sense: the attrition of scientific quality. Since the barrier for getting a scientific publication up & running is not lower: you still need staff, good editorial board, good editors, etc, etc. This all costs money, even if you do away withe the substantial expense of paper printing. But the barrier is considerably lower if you are willing to do away with credibility, and just set up what is essentially a web-based manuscript clearinghouse. Since many of those journals (like the one above) profess to be Open Access, some misinformed souls may associate Open Access ~= Bad Science. Especially if they are inundated by these emails like I am. From several publishers at that.
I worry that widespread adoption of the new policy could result in dominance of author-pays models for publication, which would squeeze out unfunded researchers from publication, and lead to a further dilution of peer review, as the journal publishers would no longer care as much about quality (no subscribers to please) as about quantity (maximizing author fees).
— Gas station without pumps, December 15, 2010
Now, bad journals, bad papers and fraud have been around for ever. In early 2009 it was revealed that Elsevier published a journal that was almost entirely ghost-written by Merck to promote Vioxx. The Elsevier fake journal and ghost writing scandal had nothing to do with Open Access. Other examples of marginal or even fraudulent publications in closed-access forums are not hard to find. Especially funny is the 2005 case of SCIgen, a program that generates random Computer Science research papers, including graphs, figures, and citations. One randomly generated paper was accepted to WMSCI 2005. (See the whole story on the scigen blog, you’ll need to page back to the beginning. This makes for a fascinating, surreal and somewhat disturbing read.)
But emails such as those above (and I get them weekly from many sources despite –or maybe because– repeated requests to unsubscribe), do show that some publishers all but say that they will publish for cash, and they obviously couldn’t care less who provides editorial and review services as long as they go through the motions of supposed peer-review. They justify the author-pays publication costs by providing Open Access — the icing on top a rather rancid cake. Unfortunately, spam such as this may cause some people to associate anything Open Access with the publication of marginal and low quality research.
Well, but if my Thunderbird email client can tell the difference between good and bad science, maybe we can educate humans about the non-association of Open Access with bad science. I jut hope that the term Open Access is not one of the parameters in my Thunderbird’s spam filter. Does anyone know how to check that?