Pawel on Open Science. Full disclosure: I consider sharing an office with this guy for over a year to be one of the best experiences of my postdoc.
She is a ‘ministering angel’ without any exaggeration in these hospitals, and as her slender form glides quietly along each corridor, every poor fellow’s face softens with gratitude at the sight of her. When all the medical officers have retired for the night and silence and darkness have settled down upon those miles of prostrate sick, she may be observed alone, with a little lamp in her hand, making her solitary rounds
— the Times newspaper, 8 February 1855
It’s Ada Lovelace Day, and time to write about your favorite woman in science.
Florence Nightingale is best known for founding the profession of modern nursing. Today nursing is a skilled degree-earning profession, requiring extensive training, with professional rights and responsibilities. That was not the case less than 120 years ago, when normally only the military and religious orders offered semi-skilled assistance to physicians. Nightingale changed all that, and revolutionizing the way medicine is practiced. Historically known as the “Lady with the Lamp”, the angel of soldiers in the Crimean War, she ministered to the wounded not only with care and compassion, but with a newly-applied professionalism. This professional approach included keeping medical records and using them to improve health care.
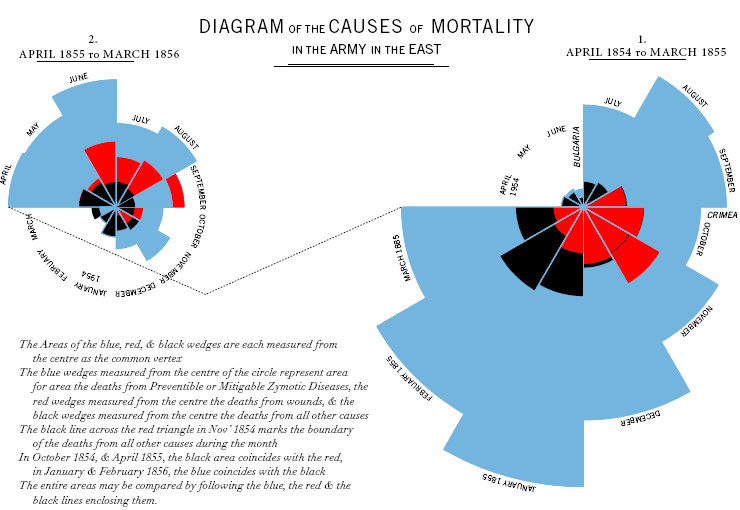
Nightingale is less known for her managerial and statistical acumen, and her pivotal role in medical statistics. Nightingale kept meticulous notes of mortality rates at the Scutari hospital in Istanbul which declined dramatically during her administration. Upon her return to London, she compiled the records into a new polar diagram, known as Nightingale Rose Chart. The data is plotted by month in 30-degree wedges. Red represents deaths by injury, blue – death by disease, and black – death by other causes.
Note that this is not a pie-chart. The wedges are all in 30-degrees (so 12 wedges/months fill a circle) and the contribution of each cause of death is proportional to each wedge’s radius. Nightingale’s visualization of the role preventable diseases play in battlefield deaths made a very strong case to military authorities, Parliament and Queen Victoria to carry out her proposed hospital reforms. Specifically for adopting hospital sanitation practices and dramatically reducing death from preventable infectious diseases.
Here is an interesting critique of the Nightingale Rose Charts which is presented at Dynamic Diagrams. It appears that by placing the preventable diseases wedge-section in the outer section of the wedge, the blue received a proportionally larger area, an artifact of this radial plot. This does not detract at all from her achievements, and, as shown in the corrected charts, not even from her case for improving hospital sanitation to reduce preventable diseases as the leading cause of death, regardless of presentation format. (Pie charts are usually problematic).
You can read more about the Mathematical affiliation of Nightingale in this excerpt form the Newsletter of the Association for Women in Mathematics. One interesting factoid: she was the first woman to be nominated a fellow of the Royal Statistical Society.
π approximation script, from Stephen Chappell.
import sys
def pi():
a, b, c, d, e, f = 1, 0, 1, 1, 3, 3
while True:
if a * 4 + b - c < c * e:
yield e
a, b, c, d, e, f = a * 10, (b - c * e) * 10, c, d, ((a * 3 + b) * 10) // c - e * 10, f
else:
a, b, c, d, e, f = a * d, (a * 2 + b) * f, c * f, d + 1, ((d * 7 + 2) * a + b * f) // (c * f), f + 2
digit = pi()
sys.stdout.write("%d.%d" % (digit.next(), digit.next()))
if len(sys.argv) < 2:
prec = 5
else:
prec = int(sys.argv[1]+1)
for i in range(prec):
sys.stdout.write("%d" % digit.next())
print
To get Pi with a precision of 10 digits:
python pi_approx.py 10
And here is another script using Machin’s formula.
I have written about the Journal of Serendipitous and Unexpected Results before and now this just popped in my inbox from JSUR’s Google group. Apparently JSUR is now open for business.
JSUR Call for Participation
Submit your short (2-4page) and full length manuscripts to the Journal
of Serendipitous and Unexpected Results.Over the past month we’ve received a great amount of press and
publicity for the Journal of Serendipitous and Unexpected Results
(JSUR). Thanks to everyone who helped spread the word, please keep it
up!In Richard Feynman’s 1966 Nobel Lecture, he said, “We have a habit in
writing articles published in scientific journals to make the work as
finished as possible, to cover up all the tracks, to not worry about
the blind alleys or describe how you had the wrong idea first, and so
on. So there isn’t any place to publish, in a dignified manner, what
you actually did in order to do the work.”We’re writing to invite you to solicit short (2-4page) and full length
submissions to JSUR. Why not prepare a 2-4 page writeup discussing
side-investigations, alleyways, or false-starts in your latest
published or unpublished research? Papers of this length place a
minimal burden on the authors, while providing extremely valuable
research insights to a broad audience.Journal website: http://www.jsur.org
Sincerely,
The JSUR Editorial Board
Yes! Why should the evolution people have all the fun with their blog carnival? (After all, it is only a theory.) It’s time for bioinformaticians to show what we are made of, and to have a carnival of our own. Bio::blogs had a good run some time ago. I decided to reconnect what is hopefully now a larger and more networked community under the title of Bioinformatics Blog Carnival.
Programming
For many new bioinformatic programmers, there is a question of which Bio* package to choose from. The Bio* packages (Biopython, BioPerl, BioJava and BioRuby) are open source licensed packages which are used heavily in bioinformatics coding. They contain parsers for bioinformatic data file, file format converters, SQL interfaces and other goodies that make a biohacker’s life manageable. Walter Jessen presents a rapid-fire no-nonsense review of Open Source Programming with Bio* Libraries on Expressing Scientific Insight. Finally, if you would like to do everything wrong, Manuel Corpas presents 10 Sarcastic Rules on How to Be a Bioinformatician posted at Manuel Corpas’ Blog.
Databases
Morgan Langille talks about BioTorrents – a file sharing resource for scientists posted at his blog Beta Science. My take on BioTorrents is that it is a cool idea, but as most institutes block BitTorrent along with other peer-to-peer sharing utilities, I doubt there is a critical mass of feeders to make BioTorrents viable. Things can change though: institutes might decide to set up dedicated servers to Torrent scientific data, just like there are legitimate Torrent servers for Linux distros. George presents Bio-graphics, BioSQL and Rails part 1 posted at Biorelated. He talks about how to quickly add graphics support to a bioinformatics database rails application.
Brad Chapman’s Blue Collar Bioinformatics is a treasure-trove of useful bioinformatics methods. Brad is very thorough in his writing, and he covers a wide variety of topics. I particularly enjoyed reading about his adventures at the biohackathon 2010 in Tokyo, and the resulting Python query interface to BioGateway SPARQL endpoint and InterMine.
Genomics
Nick Loman from Pathogens: Genes and Genomes gives some excellent tips for de-novo genome assembly. He also gives the necessary scripts,which are great companions to Velvet, the popular short read assembler. Luke Jostins writes about AGBT: Speculating on Third Gen Tech posted at Genetic Inference, “An investigation into what data from various third generation sequencing technologies may look like.” SM presents Resources for Exome sequencing annotation posted at Organizing the Strands of Curiosity.
Structural biology
Sean Seaver talks about the retraction of several protein structure papers published by one group at the University of Alabama, Birmingham. Structuregate was reported both in the media and in the blogosphere. Sean walks us through how it might have been done, and how structural bioinformatics techniques found out the wrong structures in Origin and Orientation posted at P212121. Maria Hodges talks about how difficult it is to explain structural genomics to the man in the pub. Menachem Fromer presents Tradeoff between stability and multispecificity in the design of promiscuous proteins. Promiscuous proteins are proteins which bind different partners (ligands, or other proteins). However, the more partners they are able to bind, the less stable they are, as shown in a series of simulated evolution studies, summarized at Nir London’s Macromolecular Modeling Blog. OK, promiscuity and and a pub. There must be a joke somewhere there.
A common lament among bloggers and other enthusiastic adopters of Web 2.0 technology is the lack of mainstream uptake of these tools by active scientists. A recent report from University of California Berkeley confirmed this: “The advice given to pre-tenure scholars was consistent across all fields: focus on publishing in the right venues and avoid spending too much time on public engagement, committee work, writing op-ed pieces, developing websites, blogging, and other non-traditional forms of electronic dissemination (including online course activities)“. Maria Hodges argues that Web 2.0 thrives where journals don’t, and that the NMR community might be the first to reach the tipping point, where your career is harmed by not contributing. She talks of two wikis used by the NMR community. It is a small community, which has a need for sharing methods that are generally not publishable in peer-reviewed journals. The wiki venue makes for an ideal dissemination method for this community. On the subject of data sharing, and how it can backfire: here is an interesting connection between the history of the PDB, and the email affair known as ‘climategate’.
Haiku:
A finer book of
Blog posts the world has not seen
Buy: you won’t regret
To-day we have naming of parts. Yesterday,
We had daily cleaning. And to-morrow morning,
We shall have what to do after firing. But to-day,
To-day we have naming of parts. Japonica
Glistens like coral in all of the neighboring gardens,
And to-day we have naming of parts.
— Henry Reed “Naming of Parts”
To-day we have Henry Reed’s birthday. Henry Reed, who wrote one of my favorite poems: Lessons of the War. Reed published this poem in the New Statesman and the Nation August 1942. Superficially, the poem is a soldier’s griping in the ill-equipped British army of WWII. But there is also the dark undercurrent of a man yanked from everyday life and forced to face his own mortality. Lessons of War has six parts, and it gets more somber as the poem progresses. The “naming of parts” he talks about in the excerpt above is the all-too-familiar experience in basic training: getting to know your personal weapon.
Here is part II: Judging Distances.
LESSONS OF THE WAR
II. JUDGING DISTANCES
Not only how far away, but the way that you say it
Is very important. Perhaps you may never get
The knack of judging a distance, but at least you know
How to report on a landscape: the central sector,
The right of the arc and that, which we had last Tuesday,
And at least you knowThat maps are of time, not place, so far as the army
Happens to be concerned—the reason being,
Is one which need not delay us. Again, you know
There are three kinds of tree, three only, the fir and the poplar,
And those which have bushy tops to; and lastly
That things only seem to be things.A barn is not called a barn, to put it more plainly,
Or a field in the distance, where sheep may be safely grazing.
You must never be over-sure. You must say, when reporting:
At five o’clock in the central sector is a dozen
Of what appear to be animals; whatever you do,
Don’t call the bleeders sheep.I am sure that’s quite clear; and suppose, for the sake of example,
The one at the end, asleep, endeavors to tell us
What he sees over there to the west, and how far away,
After first having come to attention. There to the west,
On the fields of summer the sun and the shadows bestow
Vestments of purple and gold.The still white dwellings are like a mirage in the heat,
And under the swaying elms a man and a woman
Lie gently together. Which is, perhaps, only to say
That there is a row of houses to the left of the arc,
And that under some poplars a pair of what appear to be humans
Appear to be loving.Well that, for an answer, is what we rightly call
Moderately satisfactory only, the reason being,
Is that two things have been omitted, and those are very important.
The human beings, now: in what direction are they,
And how far away, would you say? And do not forget
There may be dead ground in between.There may be dead ground in between; and I may not have got
The knack of judging a distance; I will only venture
A guess that perhaps between me and the apparent lovers,
(Who, incidentally, appear by now to have finished,)
At seven o’clock from the houses, is roughly a distance
Of about one year and a half.
You can read all six parts of Lessons of the War here.
I’ve been cleaning out my Gmail from attachment-emails recently. (Why do people continue to send me video files when there’s YouTube?) Anyhow, Mickey Kosloff sent me this. In this pic, form meet function, being concise and to the point, as a good grant application should be.
Byte Size Biology will be hosting the first edition of the bioinformatics blog carnival. All you bioinformatics bloggers, submit your entries by Mar 9, 2010 23:59:03 EST. Note the 3 second extension I have already given. There will be no more deadline extensions, I’ve been generous enough as it is. The carnival will be posted here by March 15, 2010.
Any blog posts that have to do with the computational aspects of: genomics, nextgen sequencing, sequence analysis, gene expression, systems biology, ontologies, databases, structural biology, metagenomics, phylogenetics, function prediction and I probably forgot a few other categories so don’t hold it against me, just submit. Early and often. Your own posts and others that you liked. Nothing too old please, 1/1/2009 and later. I reserve the right to be a less-than-benevolent dictator and screen out posts. This applies especially to commercial plugs with no other merits.
Please retweet, reblog, rebuzz and remember: submit to the bioinformatics blog carnival!
If you have a cool logo for this carnival, email bioinfo.blog.carnival AT gmail.com OhOne and OhToo will pick the winning logo to be displayed on the carnival. Likewise, if you would like to host the next edition, let me know. Do not submit posts by email, only via blogcarnival.com.
Happy carny-ing.
This has been going around the intertubes for a while now. Still very cool.
As part of the process of manufacturing a new car, the designers will take the blueprints to the factory floor. There they will set up an experimental assembly line, tinkering with the manufacturing process of the prototype until it is ready for mass-production. Can we do the same with the machinery of life — the assembly of proteins? Can we set up an alternative assembly line for a new protein prototype — and then actually set up a working assembly line for the whole new protein? A proof-of-concept has been published this week in Nature by Jason Chin’s group at the Medical Research Council Laboratory of Molecular Biology, Cambridge UK.
If there is a single common denominator to all life, it is the genetic code. All life is built around DNA encoding information for proteins nucleotide triplets or codons. Since there are four types of nucleotides (A,T,G,C) that are read in words of thee, there are 43 = 64 possible codons: more than enough to encode for the 22 amino acids that make up proteins. There is nothing more basic and fundamental to life on Earth than the three-letter based genetic code.
Until now.
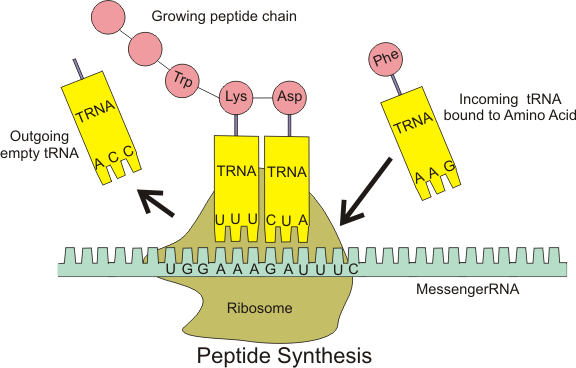
Chin’s group has created a four-nucleotide codon system. It is not that the DNA is different: it is the way the cellular machinery decoding RNA transcripts interprets the nucleotide sequence. Ribosomes –large RNA and protein complexes which are the platform upon which messenger RNA is read and decoded — are set to serve up messenger RNA three nucleotides at a time. (Messenger RNA or mRNA is a transcript of the DNA which is carried to the ribosome.) Transfer RNA or tRNA is a short RNA molecule that shuttles the proper amino acid to the ribosome, but will only attach if the proper codon is served up by the ribosome. The whole protein synthesis “assembly line” looks something like this:
To change the interpretation of the genetic code from three lettered words to four, Chin and his colleagues had to make new ribosomes, and new tRNAs. To create these new ribosomes, they designed orthogonal ribosomes, or o-ribosomes. O-ribosomes are genes inserted to produce extra ribosomes that operate in the cell alongside the regular ribosomes. The cell functions because it has the regular ribosomes to maintain its viability. The ribosomal RNA in the o-ribosomes is free to be mutated to create new unnatural traits: in this case, the ability to serve as a platform read four-letter codons. They selected for Escherichia coli bacterial cells that expressed a o-ribosomes which translated a four-letter codon in a gene, which would otherwise go untranslated by the regular ribosome. The gene gives the bacterial cells resistance to the antibiotic chloramphenicol. So cells that survive a dosage of chloramphenicol are those which have functioning o-ribosomes, as they have the chloramphenicol resistance gene that is being translated by the o-ribosomes.
They also needed to create new tRNAs that have an four-nucleotide anticodon (the part that complementarily binds to the messenger RNA — see figure above.) So the surviving E. coli cells have a population of working o-ribosomes, regular ribosomes, modified tRNA (with a four-letter anticodon) and regular tRNA.
Then they took their work a step further. Each three-letter tRNA carries a specific amino-acid, depending on its anticodon. Thus tRNAAAG will always have a phenylalanine attached, because CTT (the complement of AAG on the messenger RNA) codes for phenylalanine. If you start messing with that, the translation machinery will produce non-functional proteins, which will probably kill the cells pretty quick. But with the orthogonal 4-letter code machinery, that is not really a problem: the orthogonal machinery operates alongside the normal one. Also, there are no amino acids naturally assigned to any four letter code, because this code does not appear in nature in the first place! So Chin’s lab assigned an unnatural amino acids to a four-letter code. The non-naturally occurring p-azido-l-phenylalanine amino acid was assigned to tRNAUCCU. They then showed that the whole alternative translational machinery worked by synthesizing a mutant of the protein calmodulin which used this amino-acid in its structure.
Why do it? Well, personally I don’t see the need for justification: just being able to do it is so cool! But seriously: think of the ability to design proteins from up to 44=256 different amino acids other than the 22 we have now. The possibilities of tinkering with existing proteins using this orthogonal, four-letter based machinery are huge. The other benefit of this orthogonal synthesis setup is the ability to control this orthogonal translational machinery: because it does not use the three-letter vocabulary, this orthogonal machinery would be much easier to manipulate, tinker with and switch on and off without getting in the way of regular cellular translational machinery. The analogy to a car assembly line breaks here. It is as if two different models are being assembled on the same line just by using different robots. The better analogy is for a program source code to be read by two different compilers, each producing a different program. Awesome.
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M., & Chin, J. (2010). Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome Nature DOI: 10.1038/nature08817
Seriously, this is what I first thought when I saw the cover of this week’s Nature, and the associated drawings in the press. The dude’s haircut seems like it was even bad in the ’80s… 2080 BCE that is, which is when his body is dated. Approximately.

Inuk, artist's impression
A large group of researchers were involved in analyzing DNA from a 4,000 year old man found in Greenland. There is an interesting story on how the sample was obtained. Eske Villerslev spent 2 months looking for human remains in archaeological sites in Greenland, to no avail. When complaining about it to a friend, he learned that that friend’s father had already given hair samples to the national museum of Denmark… back in 1986. Serious slap on the forehead!
The preserved DNA was of great quality. Also, this is 2010: we have a plethora of genotypic data that lets us associate a genome with phenotypes, and geographical origin. Inuk, as he was dubbed, shares many traits with east Asians, and much less with contemporary North American Inuit or other Native Americans. This means that Inuk and his people migrated fairly recently to Greenland from Siberia and across North America. Inuk and his people, the (now extinct) Saqqaq have no relationship to contemporary Native Americans. This was also shown in an earlier study of the mitochondrial DNA.
What do we know about Inuk himself? He had blood type A+; thick black hair and very likely a receding hairline due to a baldness allele; brown eyes; shovel-graded front teeth (East Asian characteristic); dry type earwax… before you laugh that is actually, another characteristic of Asian populations on the Siberian and Chinese east coast. He also had alleles associates with human adaptation to cold temperatures.
And a bad haircut; but great press nevertheless (just got to Google News and search for “Greenland Genome” – see, for example, the NYTimes.)
 This post has been Slashdotted. Exercise extreme caution.
This post has been Slashdotted. Exercise extreme caution.
Rasmussen, M., Li, Y., Lindgreen, S., Pedersen, J., Albrechtsen, A., Moltke, I., Metspalu, M., Metspalu, E., Kivisild, T., Gupta, R., Bertalan, M., Nielsen, K., Gilbert, M., Wang, Y., Raghavan, M., Campos, P., Kamp, H., Wilson, A., Gledhill, A., Tridico, S., Bunce, M., Lorenzen, E., Binladen, J., Guo, X., Zhao, J., Zhang, X., Zhang, H., Li, Z., Chen, M., Orlando, L., Kristiansen, K., Bak, M., Tommerup, N., Bendixen, C., Pierre, T., Grønnow, B., Meldgaard, M., Andreasen, C., Fedorova, S., Osipova, L., Higham, T., Ramsey, C., Hansen, T., Nielsen, F., Crawford, M., Brunak, S., Sicheritz-Pontén, T., Villems, R., Nielsen, R., Krogh, A., Wang, J., & Willerslev, E. (2010). Ancient human genome sequence of an extinct Palaeo-Eskimo Nature, 463 (7282), 757-762 DOI: 10.1038/nature08835
Gilbert MT, Kivisild T, Grønnow B, Andersen PK, Metspalu E, Reidla M, Tamm E, Axelsson E, Götherström A, Campos PF, Rasmussen M, Metspalu M, Higham TF, Schwenninger JL, Nathan R, De Hoog CJ, Koch A, Møller LN, Andreasen C, Meldgaard M, Villems R, Bendixen C, & Willerslev E (2008). Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science (New York, N.Y.), 320 (5884), 1787-9 PMID: 18511654
If the title of this post makes you cringe, then you belong to a minority of people who realize why the phrase “highly evolved” is so wrong. Unfortunately, “highly evolved” (as an absolute term) and “more evolved” (as a comparative term) seem to be used all-too frequently. They are uttered not only by non-scientists and non-biologists but even by scientists who should know better. Even when they catch themselves after blurting out “highly evolved” in a conversation (or, more embarrassingly, in a lecture), the damage is done. Yet another Freudian (Darwinian?) slip that tells of a fundamentally bad grasp on evolution. And, yes, I know, this topic has been written about by many of my betters, who are vastly more evolved better writers than I, with much better breadth and depth of knowledge of evolution, and a reach to a much wider audience.
So why am I writing about it? Well, this is my blog and ranting in it is my prerogative. And despite the Richard Dawkinses and Steven Jay Goulds of this world, the use of this phrase still persists. So it is up to us foot soldiers of the blogging community to do our own modest bit. If I prevent any of my six readers from being tempted to utter this phrase the next time it is (wrongly) deemed appropriate, then I have done my bit.
Why is this “highly evolved” used so much? And why is it wrong?
Consider the sponge, and then consider Albert Einstein. There are certain traits that Einstein had, that a sponge does not. We deem these traits to be of merit. Einstein developed a fundamental theory in physics. He played the violin. He ate with a knife and fork, had binocular color vision, opposable thumbs and he cultivated his facial hair in the form of a mustache.
A sponge… well, to be brief, does not have all those qualities we hold in such high merit. It kinda sits there at the bottom of the shallow ocean, flopping about, filter feeding, pooping and apparently not much else. Clearly, there are qualities to Einstein that make him more interesting than the sponge.
Einstein seems, intuitively, to be more complex than a sponge, and that complexity can be quantified directly, in many ways. Actually, this is a pretty contentious point by itself: can we speak of organism complexity? Can we quantify the complexity of an organism and compare between different species? And what exactly would the complexity metric we choose tell us?
But let us assume, for argument’s sake, that our intuition that Einstein is more complex than a sponge is correct. For example, we can imagine a measure derived from the diversity and number of cells. Obviously there are more cell types in Einstein than in a sponge. Does that mean he is also more evolved? Are humans a more evolved than sponges? Chimps? After all, did life not start 3.85 billion years ago as simple and over time became more complex? Progressing, as it were from simple unicellular bacteria through more complex sponges all the way to the crowning achievement of humans? Had humans not, in a sort of (alas, Pyrrhic) victory, mastered the Earth and competed with many of earth’s species to the latter’s extinction? Isn’t competition what evolution is all about? And isn’t human victory a direct result of human complexity making humans “more evolved”? So isn’t “complexity” an end product of evolution, the more complex you are the more successful you are, and the more evolved you are?
No, no, no, no, no, and no.
The reason for this series of compounding errors is the mistaken notion that evolution by natural selection is a progression resulting in a production of increasingly complex life. Evolution is not goal oriented, and there is no teleology involved. The increasing complexity of organisms along time may seem to involve a progressive process, but there is none. It is a “statistical illusion”. What do I mean by that? Well, life did start out in less complex forms, that became more complex. But the less complex forms remained as well. Thus the distribution of complexity increased over time, but there is no directionality towards progress: the less complex life remained around as well. But over 3.85B years, complexity has had a chance to manifest itself in life, as natural selection favored some initial complexities, and those extended to become even more complex. Yes, we can trace a direct route from the first multicellular organisms, through sponges, invertebrates, vertebrates. But humans, chimps, sponges and bacteria living on Earth today are the result of exactly the same selective forces that have shaped life since it crawled out of an underwater volcano, or wherever. Complexity emerged over time, and is still emerging. But complex organisms are being added to the pool of life, rather than replacing the simple organisms. The result is an increase of a distribution of complexity levels, not the moving of an entire curve of complexity rightwards.

Apparent progress due to a to a 'wall' restricting where random change can take things. Adapted from SJ Gould. Reproduced under CC from talkorigins.org
The point I am trying to make is that humans may be more complex than sponges, but we are not “more evolved” nor are we “highly evolved”. There is no progressive process, and all of life on earth is the result of the same 3.85B years of selective pressures.
For a really good historical overview of teleological, or purpose-driven, thought in evolution, look to talkorigins.org.
All of this does not mean that Highly Evolved by The Vines is not a kick-ass song. Listening to it is also a good way to get the rage from hearing “highly evolved” out of your system. Note the low complexity of the video: