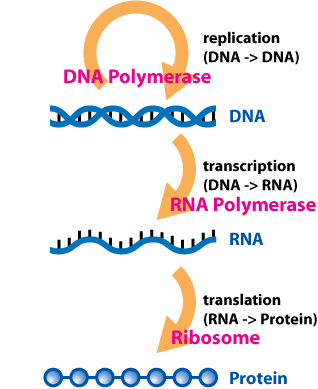
The central dogma of molecular biology edit: the sequence hypothesis (thanks for setting me straight, Kamel!) as formulated 57 years ago was simple: DNA is transcribed to mRNA,and mRNA is translated to proteins. Proteins are the business end of this process. mRNA is only the messenger: its sole function is to deliver information from the template (DNA) to the business end (Protein). It was thought to have no more function than a fax, email or instruction manual. Well, except for tRNA which carry amino acids to the ribosome. Oh, and ribosomal RNA that helps the specific binding of tRNA to the ribosomal complex, uh, yes, somehow.
This rather convenient view of RNA’s role in life has been repeatedly assaulted. The discovery that RNA can be catalytic — ribozymes — was the most dramatic change. It transformed our perception of the ribosome form a protein complex with some RNA to an active RNA complex with some protein. Since RNA was understood to both carry information and perform action, it led to the hypothesis of the RNA World. The RNA World being the primitive scaffold of molecular replicants and enzymes composed solely of RNA – later to be joined by DNA and protein. The supporters of the RNA world hypothesis see the evidence for it all around: riboswitches which are mRNA molecules that regulate their own activity by binding to small molecules; small interfering RNAs and micro RNAs are both RNA species that silence or help degrade mRNA. Small nucleolar RNA act to process tRNA and rRNA and their cousins, small nuclear RNA. All showing that the role of RNA in life is diverse, far-reaching and critical.
![]()
![]()
Last week a study published in Nature added yet another role to RNA, and from an unexpected source: pseudogenes. Pseudogenes are generally considered to be vestigial, or the remains of once active genes. The DNA sequence of a pseudogene resembles that of a gene, but owing to any of several reasons such as early termination, mutations or the inability to transcribe, pseudogenes are laying about the genome’s junkyard, accumulating rust and slowly being phased out of the genome. Until recently, pseudogenes were interesting mostly to evolutionary biologists. The reason being, since pseudogenes share ancestry with active genes, they can inform us of the evolutionary history of the genome, a sort of a molecular fossil or molecular-archaeological ruin if you will. Informing us about the past, but not having much of a function at present.
But apparently there is more to pseudogenes than just being archaeological genome dig markers, and actually they regulate the expression of their kin “real” genes. “Real” being in quotes, because suddenly pseudogenes are “real” too — they are actually doing something, not just rusting away in the genome. And performing in a very interesting way, too.
PTEN is a gene whose protein product is a tumor suppressor. In normal cells, its expression is tightly regulated, since even changes in the number of protein produced might cause cells to turn cancerous. PTEN is regulated by several types of microRNAs or miRNAs. miRNAs are non-coding RNAs, which are approximately 22 nucleotides long, and can control gene expression by interacting with their target mRNA. So miRNA molecules interact with PTEN mRNA. Each PTEN mRNA that interacts with PTEN-specific miRNA gets knocked out of circulation, lowering the number of PTEN proteins, inlreasing the chances of a cell turning cancerous. (Don’t confuse “miRNA” and “mRNA”!)
PTEN1 is a pseudogene which shares a very recent common ancestor with PTEN. A mutation in PTEN1 prevents it from being translated into a protein product, but it can still be transcribed to PTEN1 mRNA. Laura Poliseno and her colleagues have shown that PTEN1 mRNA, being very similar in sequence to PTEN mRNA attracts miRNA molecules that target PTEN mRNA. In other words, PTEN1 mRNA lures PTEN-specific miRNA molecules away from PTEN mRNA, lowering the number of inactivated PTEN mRNAs.
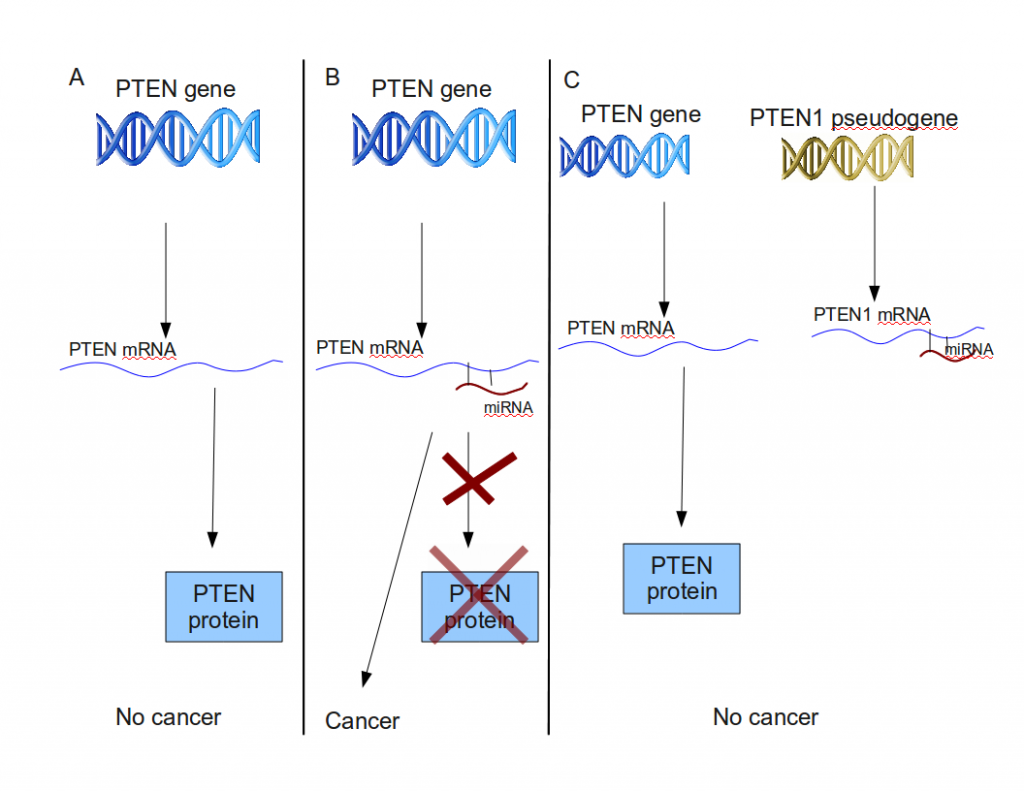
PTEN1 acts as a decoy for PTEN-specific miRNAs. (A): transcription and translation of the PTEN gene into the tumor-suppressing PTEN protein. (B): PTEN translation inhibited by miRNA. A high enough concentration of miRNA can lower the cell PTEN protein concentration below a threshold, inducing cancer. (C) The mRNA of the PTEN1 pseudogene attracts PTEN miRNA, effectively lowering its concentration in the cell, enabling PTEN protein production
So PTEN1 is not really a relic, but rather an active tumor suppression gene, on the mRNA level.
Is this PTEN/PTEN1 relationship unique? Apparently not. Looking through the mouse genome, Poliseno and colleagues have found other pseudogenes homologous to active genes, with possible mRNA binding sites. Indeed, they have shown that the same mechanism is true for the KRAS gene and the pseudogene, KRAS1. KRAS is an cancer causing gene or an oncogene, and KRAS1 acting as a miRNA decoy enhances KRAS expression. Therefore KRAS1 is also an oncogene. The authors suggest a new general model for mRNA mediated biology, which they call competitive endogenous RNA, or ceRNA.
The implications of this finding go beyond tumor suppression control. The number of pseudogenes in a typical animal genome meets or exceeds that of regular genes. It may very well be that many of them are functional on the RNA level, which offers a whole new outlook on what a gene is, what a pseudogene is, and how they function and control in the cell. It will take a while for the implication of this study to sink in, but it seems that Poliseno and her colleagues have just opened up a whole new subfield in molecular biology, with a whole new set of roles for RNA.
Oh, and if you were expecting a video of one of the most annoying songs ever, forget it.
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W., & Pandolfi, P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology Nature, 465 (7301), 1033-1038 DOI: 10.1038/nature09144