“Codon” is now a four letter word
As part of the process of manufacturing a new car, the designers will take the blueprints to the factory floor. There they will set up an experimental assembly line, tinkering with the manufacturing process of the prototype until it is ready for mass-production. Can we do the same with the machinery of life — the assembly of proteins? Can we set up an alternative assembly line for a new protein prototype — and then actually set up a working assembly line for the whole new protein? A proof-of-concept has been published this week in Nature by Jason Chin’s group at the Medical Research Council Laboratory of Molecular Biology, Cambridge UK.
If there is a single common denominator to all life, it is the genetic code. All life is built around DNA encoding information for proteins nucleotide triplets or codons. Since there are four types of nucleotides (A,T,G,C) that are read in words of thee, there are 43 = 64 possible codons: more than enough to encode for the 22 amino acids that make up proteins. There is nothing more basic and fundamental to life on Earth than the three-letter based genetic code.
Until now.
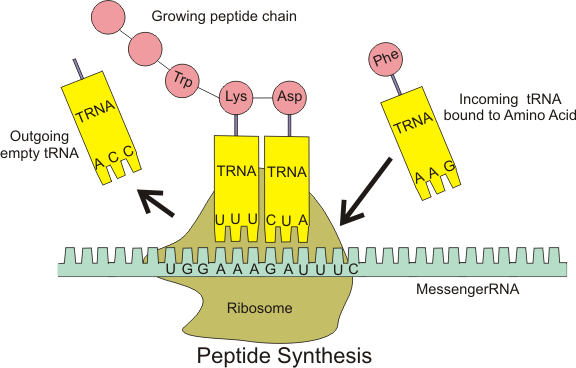
Chin’s group has created a four-nucleotide codon system. It is not that the DNA is different: it is the way the cellular machinery decoding RNA transcripts interprets the nucleotide sequence. Ribosomes –large RNA and protein complexes which are the platform upon which messenger RNA is read and decoded — are set to serve up messenger RNA three nucleotides at a time. (Messenger RNA or mRNA is a transcript of the DNA which is carried to the ribosome.) Transfer RNA or tRNA is a short RNA molecule that shuttles the proper amino acid to the ribosome, but will only attach if the proper codon is served up by the ribosome. The whole protein synthesis “assembly line” looks something like this:
To change the interpretation of the genetic code from three lettered words to four, Chin and his colleagues had to make new ribosomes, and new tRNAs. To create these new ribosomes, they designed orthogonal ribosomes, or o-ribosomes. O-ribosomes are genes inserted to produce extra ribosomes that operate in the cell alongside the regular ribosomes. The cell functions because it has the regular ribosomes to maintain its viability. The ribosomal RNA in the o-ribosomes is free to be mutated to create new unnatural traits: in this case, the ability to serve as a platform read four-letter codons. They selected for Escherichia coli bacterial cells that expressed a o-ribosomes which translated a four-letter codon in a gene, which would otherwise go untranslated by the regular ribosome. The gene gives the bacterial cells resistance to the antibiotic chloramphenicol. So cells that survive a dosage of chloramphenicol are those which have functioning o-ribosomes, as they have the chloramphenicol resistance gene that is being translated by the o-ribosomes.
They also needed to create new tRNAs that have an four-nucleotide anticodon (the part that complementarily binds to the messenger RNA — see figure above.) So the surviving E. coli cells have a population of working o-ribosomes, regular ribosomes, modified tRNA (with a four-letter anticodon) and regular tRNA.
Then they took their work a step further. Each three-letter tRNA carries a specific amino-acid, depending on its anticodon. Thus tRNAAAG will always have a phenylalanine attached, because CTT (the complement of AAG on the messenger RNA) codes for phenylalanine. If you start messing with that, the translation machinery will produce non-functional proteins, which will probably kill the cells pretty quick. But with the orthogonal 4-letter code machinery, that is not really a problem: the orthogonal machinery operates alongside the normal one. Also, there are no amino acids naturally assigned to any four letter code, because this code does not appear in nature in the first place! So Chin’s lab assigned an unnatural amino acids to a four-letter code. The non-naturally occurring p-azido-l-phenylalanine amino acid was assigned to tRNAUCCU. They then showed that the whole alternative translational machinery worked by synthesizing a mutant of the protein calmodulin which used this amino-acid in its structure.
Why do it? Well, personally I don’t see the need for justification: just being able to do it is so cool! But seriously: think of the ability to design proteins from up to 44=256 different amino acids other than the 22 we have now. The possibilities of tinkering with existing proteins using this orthogonal, four-letter based machinery are huge. The other benefit of this orthogonal synthesis setup is the ability to control this orthogonal translational machinery: because it does not use the three-letter vocabulary, this orthogonal machinery would be much easier to manipulate, tinker with and switch on and off without getting in the way of regular cellular translational machinery. The analogy to a car assembly line breaks here. It is as if two different models are being assembled on the same line just by using different robots. The better analogy is for a program source code to be read by two different compilers, each producing a different program. Awesome.
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M., & Chin, J. (2010). Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome Nature DOI: 10.1038/nature08817



















Wow…that is amazing. Could have a huge potential for synthetic biology; if you can make a ‘cell’ structure that contains synthetic DNA that codes for this synthetic ribosome, you could make all sorts of things!
I’m guessing their O-ribosomes didn’t have any ‘wobble’ usually associated with the third position of normal ribosomes.
Well, the idea of orthogonal translation is that the cell (a bacterium in this case) already encodes for the o-ribosome. Or did I miss something here?
Regarding the wobble: I am not sure what the thermodynamic aspects are. They do provide a structure of the o-ribosome in the paper. But they did not look at the thermodynamics of the 4 base codon, as far as I can tell. If anything, I am thinking there will be more wobble, as a 4-mer has more degrees of freedom than a 3-mer. Also, they only generated one working quadruple codon so far. Much more to explore.
That is really something. I am quite new to the field of synthetic biology and am trying to catch up. But even to me the implications of a four letter codon are evident! This is going to vastly enhance the repertoire of what we can do in the lab! Great post!
Very cool. Now for a new challenge … design a DNA sequence that uses overlapping 3-base and 4-base codons, where the 3-codon phase AND the 4-codon phase both produce folded protein products. This would better fit the analogy of program source code being read by two different compilers, each producing a different (but functional) program.
@Andrew Perry
That is exactly what they did with the calmodulin. One tetracodon was read in-frame to add the p-azido-l-phenylalanine to the calmodulin. the rest of the protein was translated from tricodons.