Protein Function: how do we know that we know what we know?
The trouble with genomic sequencing, is that it is too cheap. Anyone that has a bit of extra cash laying around, you can scrape the bugs off your windshield, sequence them, and write a paper. Seriously?
Yes, seriously now: as we sequence more and more genomes, our annotation tools cannot keep up with them. It’s like unearthing thousands of books at some vast archaeological dig of an ancient library, but being able to read only a few pages here and there. Simply put: what do all these genes do? The gap between what we do know and what we do not know is constantly growing. We are unearthing more and more books (genomes) at an ever-increasing pace, but we cannot keep up with the influx of new and strange words (genes) of this ancient language. Many genes are being tested for their function experimentally in laboratories. But the number of genes whose function we are determining using experiments is but a drop in the ocean compared to the number of genes we have sequenced and whose whose function is not known We may be sitting on the next drug target for cancer or Alzheimer’s disease, but those proteins are labeled as “unknown function” in the databases.
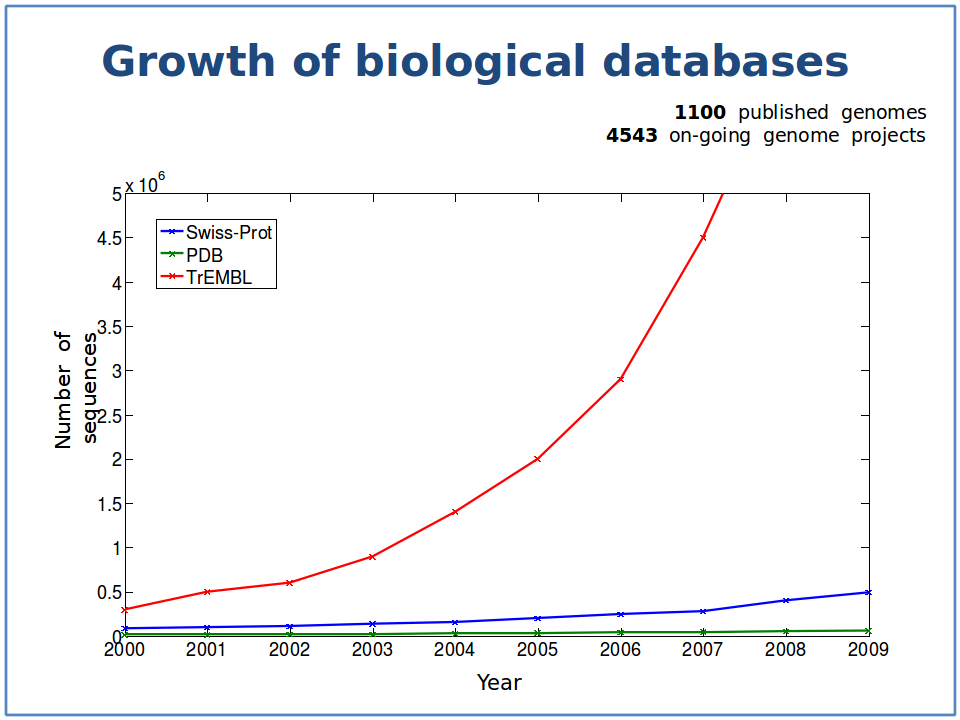
The red line is the growth of protein sequences deposited in TrEMBL, a comprehensive protein sequence database. The blue line illustrates the growth proteins in TrEMBL whose function is know, or at least can be predicted with some reasonable accuracy. The green line is the growth in the proteins whose 3D structure has been solved. Note the logarithmically increasing gap between what we know (blue) and what we do not know (red). Image courtesy of Predrag Radivojac.
Enter bioinformatics. CPU hours are cheaper than high throughput screening assays. And if the algorithms are good, software can do the work of determining function much cheaper than experiments. But therein lies the rub: how do we know how well function prediction algorithms perform? How do we compare their accuracy? Which method performs best, and are different methods better for different types of function predictions? This is important because most of the functional annotations in the databases come from bioinformatic prediction tools, not from experimental evidence. We need to know how accurate these tools are. Think about it this way: even an increase of 1% in accuracy would means that hundreds of thousands of sequence database entries are better annotated, which in turn means a lot less time in the lab or in high throughput screening labs going after false drug leads.
So a few of us got together and decided to run an experiment to compare the performance of different function prediction software tools. We call our initiative the CAFA challenge: Critical Assessment of Function Annotation. There are many research groups that are developing algorithms for gene and protein function prediction, but those have not been compared on a large scale, yet. OK then: let’s have some fun. We, the CAFA challenge organizers, will release the sequences of some 50,000 proteins whose functions are unknown. The various research groups will predict their functions using their own software. By January 2011 all the predictions should be submitted to the CAFA experiment website. Over the net few months, some of these proteins will get annotated experimentally. Not many, probably no more than a few hundred judging by the slow growth of the experimental annotations in the databases. But we don’t need that many to score the predictions. A few dozen will do.
On July 15, 2011 we will all meet in Vienna, and hold the first-ever CAFA meeting as a satellite meeting of ISMB 2011. This will be the fifth Automated Function Prediction meeting we have been holding since 2005. Only this time, there won’t just be the usual talks and posters, there will be the results of a very interesting experiment. The International Society for Computational Biology is generously hosting our meeting, and judging by the response we are getting so far, we will need one of the larger halls.
Learn more at http://biofunctionprediction.org If computational protein function prediction is your thing, join the CAFA challenge. If you are just an interested observer, keep an eye on the site. In any case, please spread the word. Finally, if your company wants some publicity, get in touch! We could use the sponsorship ^_^
Acknowledgements: I would like to thank the CAFA co-organizers, Michal Linial and Predrag Radivojac. The CAFA steeering committee: Burkhard Rost, Steven Brenner, Patsy Babbitt and Christine Orengo for supporting us, keeping us on the straight and narrow and for incredibly useful and insightful suggestions. Sean Mooney and Amos Bairoch for hashing out the assessment. Tal Ronnen-Oron and the rest of Sean Mooney’s group for setting up the CAFA website. The International Society for Computational Biology for sponsoring us. The community of computational function predictors that have participated in and supported past meetings on computational function prediction, the research groups that have registered to CAFA so far, and those that will register soon 🙂 Finally, Inbal Halperin-Landsberg for coining the name CAFA. I apologize in advance if I left someone out.
GO CAFA!
Godzik, A., Jambon, M., & Friedberg, I. (2007). Computational protein function prediction: Are we making progress? Cellular and Molecular Life Sciences, 64 (19-20), 2505-2511 DOI: 10.1007/s00018-007-7211-y




















Shouldn’t that be an ‘exponentially increasing gap’ in the figure caption?
Fantastic! Exactly what’s needed.
Are there any plans to deal with the multifunction aspect of proteins? I’m thinking in terms of the “experimentally” verified?
Looks like a really good effort.
@cement_head
We are using GO as the annotation standard. If a protein is found to have more than one GO term associated with it, then predicting the a subset or the full complement of GO terms will raise the prediction score. The rules are available here: http://biofunctionprediction.org/node/262
cement_head brings up an interesting point. Computational methods can detect multiple functions in a protein in parallel, while experimental verification of function would presumably be done serially (if multiple experiments are even done, once an initial function is discovered) and so the additional functions would take much longer to be identified. We have to start assessing somewhere, but some “wrong” predictions may simply be “ahead of their time”.
Are there ways people use to probe protein function experimentally that allow for identification of multiple functions in a single set of experiments (without strict hypotheses about what those functions might be)?
@shwu
Well, the most obvious is ligand-binding assays can detect multiple affinities (at different binding constants) of different ligands. This does not necessarily mean that all the ligands bound are physiological, and sometimes that does not even matter e.g. in the case of testing for a good pharmaceutical binder. So we may characterize a protein as “ATP binder” but also as “caffeine binder” (per example).
Taking it one step further, high throughput catalytic assays can check for catalysis of said ligands, at different Kcat constants. Again, checking only for an enzymatic aspect of function, without previous hypotheses.
But the same assays cannot be used to, say, test for physiological functions. Y2h or a protein array be used for discovering protein-protein interactions. This can inform us of the physiological aspects — again, no strict hypotheses, but usually no deep conclusions either.