Gradualism is a cornerstone principle in evolution. Things happen in small increments; all the time that changes happen, overall organism fitness cannot be compromised. So how then do full-featured complex functions appear? One way is by functional re-purposing of an existing organ:
…an organ may become rudimentary for its proper purpose, and be used for a distinct object: in certain fish the swim-bladder seems to be rudimentary for its proper function of giving buoyancy, but has become converted into a nascent breathing organ or lung. Other similar instances could be given.
— Charles Darwin Origin Ch. 13
A similar re-purposing phenomenon happens in enzymes, and the mechanism by which it appears was dubbed
enzyme promiscuity. The idea behind enzyme promiscuity is simple: an enzyme E may catalyze a reaction S1 –>P1
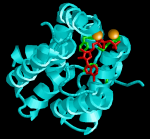
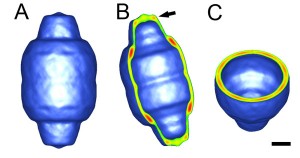
C. freundii DHAK structure. Cyan: ATP binding domain. Red: ATP. Green: ATP binding residues on the protein. Orange spheres: Mg2+ atoms. Click for larger image.
(substrate 1 to product 1) with a certain degree of efficiency. However, E may also catalyze S2 –> P2 albeit with much lesser efficiency. Now suppose the gene for this enzyme E gets duplicated in the genome, not an uncommon occurrence: one gene copy E1 goes ahead through the generations catalyzing the original reaction S1–>P1. The second copy, E2, is now superfluous for performing S1–>P1. E2 is now “free to evolve”, with no fitness constraints tying it to the original function. If the new reaction S2–>P2 increases the organism’s fitness, then E2 is under positive selection, for mutations increasing its ability to create P2 from S2. For example, P2 may be a rare nutrient, but S2 a plentiful one, thus any organism that can make its own P2 is at an advantage in S2-poor environments.
In a very elegant work published this week in Chembiochem, Eduardo Jucenda and his colleagues have captured a snapshot of the evolution of enzyme promiscuity, with the old function maintained, the new one evolving, and without gene duplication necessary. The”E1″ discussed is dihydroxyacetone kinase or DHAK, which the group has been researching in the bacterium C. freundii. Like many enzymes, DHAK uses a metal ion as a cofactor to perform its catalysis, in this case magnesium (Mg2+). During that time another group, Cameselle and co-workers have reported that another enzyme, FMN cyclase isolated from humans, is functionally promiscuous: it catalyzes both a DHAK reaction and an FMN cyclase reaction. FMN cyclase uses manganese ion (Mn2+) as a cofactor. Since the sequence of the human FMN cyclase has a 40% sequence identity with their bacterial DHAK, Jucenda’s group have decided to investigate if the converse is true: whether the bacterial DHAK can catalyze an FMN-cyclase reaction. They discovered that the bacterial DHAK can perform FMN-cyclase activity, but it needed Mn2+ as a cofactor (remember: bacterial DHAK uses Mg2+). Furthermore,when going above a certain Mn2+ concentration, the kinase activity is inhibited, while the cyclase activity is not (although the cyclase activity in the bacterial enzyme is much weaker than in the rat enzyme). So what Jucenda’s group has discovered was not just a promiscuity event, but a way to switch between two different activities by changing the concentration of Mn2+ in the reaction. This switch promotes activity 2 (cyclase) while maintaining activity 1 (DHAK), in the absence of gene duplication, this mechanism can maintain both activities. C. Freundii can actually raise it’s Mn2+ concentration temporarily, which supports the hypothesis that the researchers have discovered a new mechanism for switching between cyclase and kinase activities, and its mechanism of evolution.
Further reading:
Israel Sánchez-Moreno, Laura Iturrate, Rocio Martín-Hoyos, María Luisa Jimeno, Montaña Mena, Agatha Bastida, Eduardo García-Junceda (2009). From Kinase to Cyclase: An Unusual Example of Catalytic Promiscuity Modulated by Metal Switching ChemBioChem, 10 (2), 225-229 DOI: 10.1002/cbic.200800573
A very interesting review on enzyme promiscuity:
O KHERSONSKY, C ROODVELDT, D TAWFIK (2006). Enzyme promiscuity: evolutionary and mechanistic aspects Current Opinion in Chemical Biology, 10 (5), 498-508 DOI: 10.1016/j.cbpa.2006.08.011