The polypharmacome
![]()
![]()
Pharmaceutical companies are always on the lookout for secondary drug targets. After all, if you invest billions developing a single drug, you would be more than happy to sell it as a treatment for two, three, or more different ailments. Sildenafil citrate was developed to treat angina and hypertension, but during phase I clinical trials, it was found that Sildenafil induces penile erections. The drug was branded Viagra, and the rest is history. Eflornithine, an anti-cancer drug, is also effective against the agent of African sleeping sickness, Trypanosoma brucei. African Sleeping Sickness is known as a “neglected disease”, for which drug development is not profitable and therefore not a priority. However, having a drug already on hand makes it easier to distribute in affected areas, since the R&D costs are recovered elsewhere.
Another example of polypharmacology is a drug that binds to multiple targets in the human body. This could be used for overcoming drug resistance, a known problem with cancer. Cancer tumors often develop a resistance to anti-cancer drugs by simple natural selection: the protein that the drug binds to mutates, and no longer binds the drug. However, if the drug acts by binding redundantly to several proteins, it would be more effective, since several mutations would be required to effect drug resistance.
Another important polypharmacological consideration is toxicity. If a drug binds to one protein, its drug target, it may also bind to another one which it should not bind to as it disrupts the normal functions and the health of the patient. If the side effects outweigh the cure, the drug is no good.
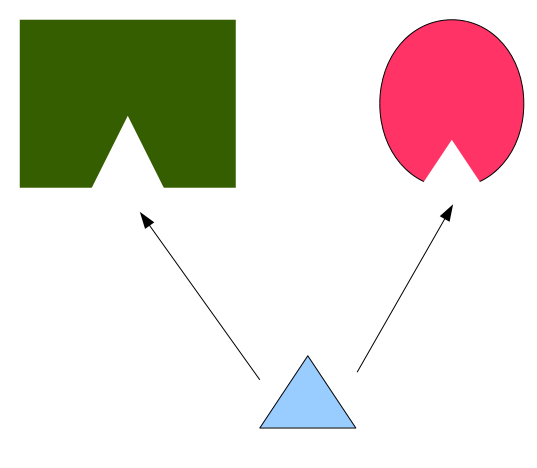
How one drug (cyan) can bind to two different proteins with different overall shapes (pink and green), but with similar binding sites
Because it can either increase profits, or conversely derail a whole process of drug development, predicting polypharmacophoric effects is very much something drug developers want. A study published yesterday in PLoS Computational Biology by Jacob Durrant and his colleagues suggest a way bioinformatics and theoretical biophysics can help in identifying multiple drug targets. Durrant’s goal was simple: given the molecular structure of a candidate drug, which proteins are expected to bind it? The strategy this group took is a combination of bioinformatic and experimental screening.
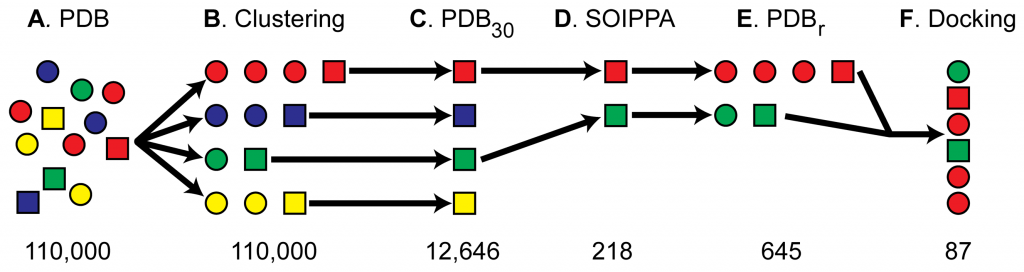
Finding additional drug targets in four steps
Step 1 (A-C in the figure above): identify the known target protein. Now pick all the protein structures that are not similar to it. Why those that are not similar? Similar proteins could be potential drug targets, since they have a similar shape to the known target protein. But here they are interested in finding targets from proteins that are of a different shape, and have no homology to the known target protein: secondary targets.
Step 2 (D): take this set of dissimilar proteins, and look for binding site similarities. Binding sites are clefts in the protein that may bind drugs. If those clefts are similar in shape to the cleft in the known target proteins, they may bind the drug. Leave only those non-homologous proteins that have similar binding sites (D in the figure)
Step 3 (E): Now add all the proteins that are homologous to the set generated in step 2. This increases the number of possible targets to homologs of the proteins that were initially selected only for binding site similarity.
Step 4 (F): take the drug molecule, and try to dock it to the various protein structures. Rank the druggability of each protein according to the score provided by the drug docking software (Autodock).
Experimental verification
Now for the cool part. Durrant and colleagues tested the computational prediction in the lab, using the compound NSC-45208. NSC-45208 inhibits a protein responsible for RNA processing in Trypanosoma brucei. So we know it is a potential drug against African sleeping sickness. What else is it good for?
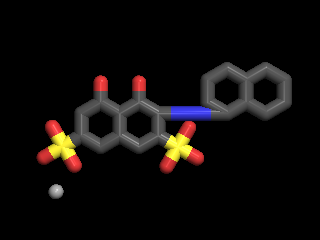
(NSC-45208), 4,5-dihydroxy-3-(1-naphthyldiazenyl)-2,7 -naphthalenedisulfonic acid, a recently discovered inhibitor of T. brucei RNA editing ligase 1 (TbREL1)
“The predicted secondary targets that gave the best docking scores, H. sapiens mitochondrial 2-enoyl thioester reductase (HsETR1), T. brucei UDP-galactose 4′ epimerase (TbGalE), H. sapiens phosphodiesterase 9A (HsPDE9A2), and Streptococcus pneumoniae teichoic acid phosphorylcholine esterase (SpPce), were subsequently tested experimentally.”
Durrant and his colleagues tested their predictions that NSC-45208 also binds to two human proteins (HsETR1 and HsPDE9A2), one additional Trypansome protein (TbGalE), and a bacterial (Streptococcus pneumoniae) protein (SpPce). Not only binds, but also inhibits their enzymatic activity. Their predictions worked well on the top two predicted targets: NSC-45308 inhibited the enzymatic activity of HsETR1and TbGalE, but HsPDE9A2 and SpPce were not affected by the drug.
At first blush, this does not seem to be much of a batting average: two positives and two false positives. But we have to remember that the experimental verification of these predictions — even if you have a good enzymatic assay to check predictions– can be very time- and resource consuming. An exhaustive test of the predictions for several compounds and many target enzymes is still not possible. As initial proof-of-principle, this work goes much farther than most other joint experimental / computational works I have read. The authors also go into lengthy and detailed discussions on limitations and improvements, as well as on another form of non-specific inhibition of the secondary targets, makes for a really interesting read on polypharmacological considerations in drug screening.
Durrant, J., Amaro, R., Xie, L., Urbaniak, M., Ferguson, M., Haapalainen, A., Chen, Z., Di Guilmi, A., Wunder, F., Bourne, P., & McCammon, J. (2010). A Multidimensional Strategy to Detect Polypharmacological Targets in the Absence of Structural and Sequence Homology PLoS Computational Biology, 6 (1) DOI: 10.1371/journal.pcbi.1000648



















Thanks for your post about this article. Very interesting! I recently came across another interesting article entitled “Predicting new molecular targets for known drugs” http://docking.org/?q=node/152