Life Stands on the shoulders of Giants (Viruses)
Back to ancient life, what exactly defines life, and where does life end and non-life begin. One of my favorite subjects, and one of which I am the least knowledgeable. Doesn’t stop me writing about it though.
Viruses are… well… not really life. Or so says common wisdom. They have some elements of life: a genome, the ability to reproduce, and being subject to evolution by natural selection. But they cannot reproduce independently: they need to hijack the reproductive machinery of an actual living cell to do that. They do not have a metabolism: they are basically syringes with DNA or RNA, equipped with basic sensors that help them lock onto cells and use them to reproduce, usually destroying their hosts in the process. One thought is that viruses evolved as segments of DNA and RNA that managed to mobilize themselves between different cells. This is the escape hypothesis of viral evolution. Evidence for this hypothesis, at least for certain types of viruses, lies with the very small number of genes many viruses have: HIV only has 4 genes (or more like 10, depends on how you count). HIV and similar retroviruses appears to originate from mobile RNA elements or retrotransposons, which use a small number of genes to replicate themselves within genomes. Indeed, it is estimated that 42% of the human genome is composed of some kind of retrotransposon, and in wheat retrotransposons constitute 90%(!) of the genome. Retroviruses like HIV may be retrotransposons that managed to escape the confines of a single organism, using a rudimentary protein vehicle to transport themselves. They still replicate by integrating into the genome of its host.
Serious cracks in the dominance of the escape hypothesis came with the discovery of the Mimivirus (see also this post). Mimivirus – Microbial mimicry virus was actually mistaken for a bacteria for nearly a decade after it was discovered, hence the name. The Mimivirus is as large as a bacterium and its genome has 979 genes — there are actually bacteria out there (albeit parasitic), with fewer genes. Some of the Mimivirus genes seem unnecessary, and 10% of its genome does not even code for genes: that is a very large chunk of seemingly wasted genome for viruses, who generally exploit all their genomic real estate very efficiently. But when your capsid has a volume close to that of a bacterial cell, then natural selection probably doesn’t pressure you that much to skimp on every nucleotide. Other giant viruses are the Mamavirus, Cafeteria roenbergensis (ya, rly), and Megavirus, the largest virus known to date with a whopping arsenal of 1,120 genes. All with large genomes, all with genes that are not found in other viruses, and seem somewhat unnecessary in a virus. Including some genes needed for translation. All this lead to a revival of an hypothesis first raised in 1924 but which has not caught on: the regressive hypothesis. The regressive hypothesis states that viruses evolved from a living ancestor that was parasitic, and through cumulative gene-loss eventually crossed the line to “non-life”. We see evidence of genome loss in parasitic bacteria: Rickettsia is a parasitic bacterium, which it has evolved from free-living ancestors. Rickettsia and mitochondria, intracellular organlles with only the remnants of a genome, share a common, free-living ancestor. It may very well be that giant viruses have evolved from more complex, organisms that lost genetic information over time, becoming parasitic and eventually non-living (or undead, if you like).
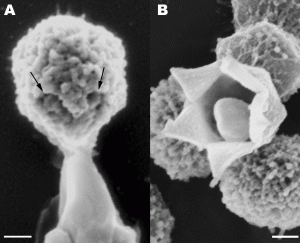
Stargate Opening From: Distinct DNA Exit and Packaging Portals in the Virus Acanthamoeba polyphaga mimivirus Zauberman N, Mutsafi Y et al. (2008) PLoS Biology Vol. 6, No. 5, e114 doi:10.1371/journal.pbio.0060114
While it is fairly accpeted that HIV’s evolution fits the escape hypothesis, there is a debate about the origins of giant viruses. Some argue that giant viruses may be the evolution of a parasite into a virus, and that the ancestors of giant viruses were once bona-fide living organisms, with metabolism, homeostasis, autonomous replication and all that jazz. Both the escape and regressive hypotheses may be true, but they simply fit different groups of viruses.
One study which supports the regressive hypothesis for giant viruses was recently published by a group from the University of Illinois, Urbana-Champaign in BMC Evolutionary Biology. The authors used protein structures, rather than DNA sequences or protein sequences to determine evolutionary relationships between archaea, bacteria, eukarya and giant viruses. There are considerably fewer protein structural templates (also called “folds”) than protein sequences. In other words, life has evolved only a limited number of structures necessary to conduct its business, although there are many sequences than map into the relatively small number of folds (an estimated 1,000-10,000 folds). The rationale of Nasir and colleagues was, that if we look at the distribution of protein structures in life’s superkingdoms, we may be able to tell whether giant viruses form an ancient separate domain. Or, as the authors call it, a supergroup, since like the Traveling Wilburys, giant viruses may have been alive once, but no longer. Therefore viruses do not merit the classification of domain, which is reserved for living things.
The authors show that, based on the presence or absence of protein Fold Superfamilies (FsF, a level of structural classification of proteins), viruses do indeed form a separate supergroup, and may have been a separate supergroup, predating the Last Universal Common Ancestor, or LUCA.
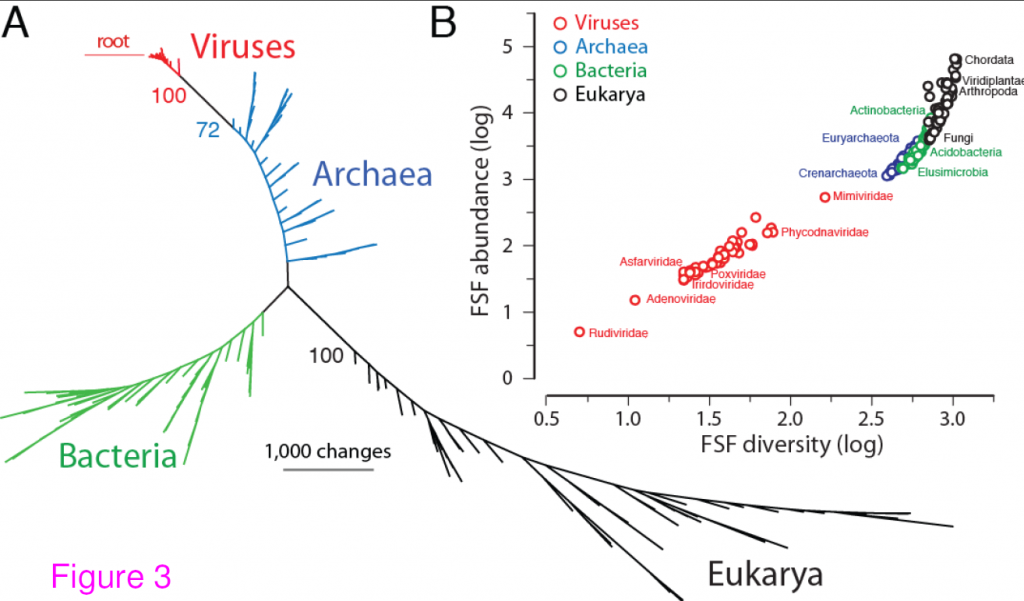
Universal tree of life (uToL) and proteomic diversity. A. One optimal (P < 0.01) most parsimonious phylogenomic tree describing the evolution of 200 proteomes (50 each from Archaea, Bacteria, and Eukarya and viruses; virus families are listed in Table 2) generated using the census of abundance of 1,739 FSFs (1,517 parsimoniously informative sites; 62,061 steps; CI = 0.156; RI = 0.804; g1 = −0.325). Terminal leaves of Viruses (V),Archaea (A), Eukarya (E) and Bacteria (B) were labeled in red, blue, black and green respectively. Numbers on the branches indicate bootstrap values. B. FSF diversity (number of distinct FSFs in a proteome) plotted against FSF abundance (total number of FSFs that are encoded) for 200 proteomes. Major families/phyla/kingdoms are labeled. Both axes are in logarithmic scale. Reproduced from BMC Evol Biol. 2012 Aug 24;12(1):156 under CC-BY license
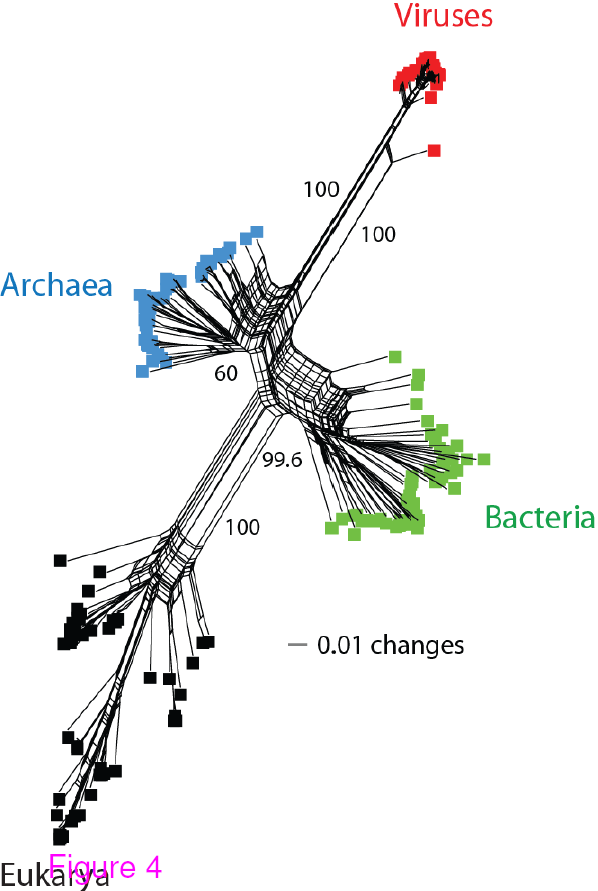
Network tree visualization of the supergroups. Network tree generated from the
presence/absence matrix of 1,739 FSFs in 200 proteomes sampled equally from the four
supergroups. The number of non-constant sites was 1,581. Nodes in the network tree are proteomes and are represented by rectangles labelled red, blue, green, and black for viruses, Archaea, Bacteria and Eukarya, respectively. Numbers on the major splits indicate bootstrap values. Reproduced from BMC Evol Biol. 2012 Aug 24;12(1):156 under CC-BY license
The authors conclude:
We show that viruses with medium-to-very large proteomes harbor a significant number of
FSFs and suggest that they have evolved via massive reductive evolutionary processes that
are not so uncommon for small bacteria with small genomes and similar parasitic lifestyles.
In addition, these viruses appear as a distinct supergroup on a uToL along with the three
cellular superkingdoms. We propose that the viruses we have analyzed coexisted with
primordial cells. These primordial entities could have been integral components of the
common ancestor of life.
I found their results to be quite compelling, at least in terms of FsF distribution among the supergroups. I wonder what would happen with an analysis that includes more than just NCLDVs (Nucelocytoplasmic Large DNA viruses, the viruses mainly used in this study). I would like to see how much further this distinction in protein fold distribution remains between viruses and cellular life, and when it starts to blur. I suspect that for many of the smalller viruses, they would be more like their hosts than like each other, if they were formed by genome escape, rather than by reductive evolution. Another good followup to this study would be to look at much larger groups of viruses, and see if other cluster into supergroups based on FsF distributions in their genomes. Because if too many different groups of viruses would cluster into distinct supergoups based on the distribution of protein structures, this may indicate that using FsFs may not be a good phylogenetic marker for this purpose. But as it looks so far, the hypothesis that at least some extant groups of viruses may have formed an ancient domain of life is definitely worth exploring further.
Read more on the origin of viruses.
Nasir, A., Kim, K., & Caetano-Anolles, G. (2012). Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya BMC Evolutionary Biology, 12 (1) DOI: 10.1186/1471-2148-12-156



















[…] Life Stands on the shoulders of Giants (Viruses) (bytesizebio.net) […]
[…] Life Stands on the shoulders of Giants (Viruses) (bytesizebio.net) […]