DIGging into Images and Genomes
Our lab has a new project and website up. The project is BioDIG: Biological Database of Images and Genomes. BioDIG lets you combine image data and genome data of, well, just about anything which you can make images and have a genome, or partial genomic information.
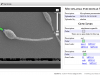
You can upload your image, annotate (tag) parts of it, and associate the tags with one or more genes. Clicking on the tags will take you to a genome browser centered on the genomic region of interest. You can use this, for example, to take cellular images and point out specific areas of protein expression. Or cell pathologies associated with mutations. Or your favorite knockout mouse… anything that has a phenotype and an associated genoype. BioDIG allows for collaborative work, so collaborators can work together on images.
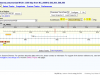
An example, very much in development database is here: MyDIG, the Mycoplasma Database of Images and Genomes. Go here for an example of a tagged image that leads you to the genome browser. Or browse through the screenshots:
For the the genome browser we used GMOD (Generic Model Organism Database) technologies. The genome browser is GBrowse; the genome database is Chado + a few hacks of our own (like an SQLite layer that gets things done faster). This is a good opportunity to thank Scott Cain and all the people of the GMOD community who have been helping us.
BioDIG is very much work in progress, and when I say “we” I mean my graduate student, Andy Oberlin, who developed and coded, and my programmer, Rajeswari Swaminathan: I’m just trying to keep up with them. Right now we have a paper in review, and a preprint on Arxiv.org.
BioDIG is now mature enough to start getting more programmers and users involved. So if you are interested in linking images to genomes, or , more generally, genotypes to phenotypes, please visit biodig.org. The code is up on our github repository, under and open source license. Feel free to fork, and get busy helping. We have a whole list of todos and a development plan. We also love to hear new ideas. Contact Andy, Raji, or myself if interested in contributing, or in setting up your own BioDIG-enabled website.





















Wow! Nice! Although (of course I would say this…) you should use jbrowse – it is ideal, would be slicker/faster/better, and is compatible with GBrowse in lots of ways.
A great idea. What is the image store? Have you seen the open microscopy environment? It’s more than just microscopy.
Ian: we wanted to use JBrowse, but a couple of things we needed (or will need in the future) were not yet implemented. It is slick & fast, and we liked the client side aspect which takes the load off the server. Hopefully soon.
David: didn’t know about the OME, thanks. We are just using a simple MySQL and image files to run the image module, but we’ll look into this.
Ian: To clarify what Iddo said we chose GBrowse because at the time we implemented this system (and correct me if I am wrong) JBrowse did not have the search feature that GBrowse has, which is important for our application.