It’s a smORF world, after all?
Here is a study that looked for a type of genes that the authors felt was neglected by classic genomic annotation. The research shows how to employed concepts in molecular evolution to validate the existence of these genes.
Some background: the first question we ask after assembling a genome is: “where are the genes”? Not an easy question to answer, since a gene is classically defined as a unit of heredity. It may code for RNA, protein, or sometimes, nothing at all. The actual implementation of the “unit of heredity” can take several physical forms, each one of them different. Therefore, the algorithms for finding genes would depend on which type gene one is looking for, exactly.
A somewhat more tractable question is: “where are the open reading frames”? Open reading frames or ORFs are those stretches of DNA that code for proteins. Indeed, most gene calling software actually identifies ORFs. There are many attributes that go into an ORF calling algorithm: the frequency of the bases (or k-mers of bases) in the suspected coding regions, the signals for the beginning and ends of introns, the existence of non-coding regions that aid transcription such as promoters and enhancers, the location on the chromosome with relation to other ORFs, and the length of the of the final product. The latter criterion is actually quite important, as many ORF-calling algorithms will discount anything coding for a protein that is shorter than 100 amino acids as being “too short”. The reason for employing this length cutoff, is that the number of false positives increases dramatically when ORFs coding for proteins shorter than 100aa (or 300 nucleotides) are called. Therefore, most gene-callers would just tend to discard any short peptides.
But throwing away the baby with the bathwater is not a good solution, since short peptides are known to be responsible for many of life’s activities: mating pheromones, small compound transporters, hormones, neurotransmitters and regulation of other proteins’ activities, to name a few. Many of these short peptides are the result of the cleavage of larger proteins, which means that the ORFs encoding for them are originally longer than 300bp. But some may actually have their own ORFs, coding only for them. How can we find those small ORFs or smORFs out? How many of them are there? Is the number of smORFs large enough to make it worth re-annotating genomes?
Emmanuel Ladoukakis from the University of Crete and colleagues from the university of Essex, UK have set up a bioinformatic pipeline to look for smORFs in the Drosophila melanogaster genome. Bear with me, there are a few steps in this pipeline. But there’s a lot to learn about genomics just from looking at what they did, and why they took those steps.
Here’s what they did: 1) Find smORF candidates: they looked for all potential smORFs (starting with a start codon and ending with an in-frame stop codon, 30-300bp long) in those parts of D. melanogaster’s genome that were annotated as non-coding. To keep things simple, they looked only for intron-less smORFs: smORFs that are encoded consecutively in the DNA. They found 593,586 potential sequences. 2) Remove transposons: they then removed all those that had a similarity to transposons. Transposons are DNA elements that multiply in the chromosome: something like an internal virus, only usually benign. They may carry bits of other genes they “grab” on the way, but they are not functional. They were left with 556,554 sequences 3) Big step: look for homologs in another fly species: they then looked for smORFs with similar translated amino-acid sequences in D. pseudoobscura, which diverged from the melanogaster 25 to 55 million years ago. The reason they looked for similar amino-acid sequences was that if there is a selection to conserve a smORF, it would be on the protein, and not at the DNA level. This step reduced the number of smORF candidates by 93%: from 556,554 down to 43,210. Looking only for 4) global alignments, (another big step) they found 4,561 smORF candidates by looking at alignments of whole smORF sequences, not only of partial local similarities. this reduced the number of candidates by 72% from the step (3). We are now down to 0.8% of the original 593,586 smORF candidates.
Quite a filtering process. Note the huge elimination: 99.2% of all initial smORFs candidates are gone. I believe that they decided to sacrifice sensitivity in favor of specificity
So they had 4,561 smORF candidates conserved between two flies. Still, how many ORFs got in by chance? Hard to know, but they continued to rely on evolutionary conservation as a guideline. There may be smORFs that appeared independently in melanogaster and pseudoobscura after they separated 55 million years ago, but the main evidence for true smORFs would be their evolutionary conservation between the two fly species.
To get even more specific, they now 5) looked for shared synteny: conservation not only of sequence, but also of the genomic context: the sequences surrounding it. That brought the number down to 3,314.
OK, so they looked for conservation based on homology and based on synteny. Anything more? Well, yes. The next step would be to 6) look for evolutionarily selected smORFs. The two evolutionary criteria they used until now were homology and synteny. Now comes a third: selection. If smORF candidates are actually coding, they will be subject to purifying selection, that is, to selection that eliminates deleterious mutations. This is evident in a low rate of non-synonymous vs. synonymous substitutions, or a Ka/Ks ratio of << 1. (Read about Ka/Ks ratios also here.) 7) Looking at what actually gets transcribed in Drosophila (from looking at the transcriptome) this number was whittled down to a final 401.
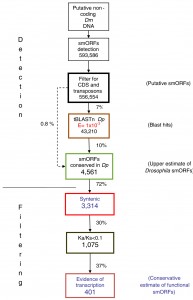
Click to enlarge. Search pipeline for Drosophila smORFs. Diagram of the smORF search pipeline followed in this study. The percentages of smORFs passing each filter are indicated. For full details, see Results and Materials and methods. CDS, coding DNA sequence; Dm, Drosophila melanogaster; Dp, Drosophila pseudoobscura; Ka/Ks, ratio of non-synonymous (Ka) to synonymous (Ks) nucleotide substitution.Ladoukakis et al. Genome Biology 2011 12:R118 doi:10.1186/gb-2011-12-11-r118
So the chosen 401 smORFs are evolutionarily conserved, both in sequence and in synteny, subject to purifyng selection (by Ka/Ks ratio) and produce a transcript. The authors obviously went for specificity over sensitivity: they looked for “good bet” smORFs rather than a large number of candidates. What I like about this study is the way that the authors used a large number of evolutionary traits that can be used as attributes for identifying smORFs. They also were careful to rule out, as much as possible, that these smORFs that may be a result of a larger transcript. This is a really nice molecular evolution work. There is no experimental evidence yet of the functionality of these smORFs: those are left to future proteomic and fly geneticists. But the idea of a small(er) world of genes, hiding in plain site among the more familiar large ones, does have its appeal, and may yield some surprises about how are genomes are structured.
Finally, for the evolutionary biologists: read the paper; there is quite a lot more to it that what I wrote. I just gave the highlights.
Ladoukakis, E., Pereira, V., Magny, E., Eyre-Walker, A., & Couso, J. (2011). Hundreds of putatively functional small open reading frames in Drosophila Genome Biology, 12 (11) DOI: 10.1186/gb-2011-12-11-r118
http://genomebiology.com/2011/12/11/R118/abstract



















Thanks a lot for this. It’s an important and controversial subject; especially in relation to the extent of lincRNAs. I especially liked the finding that tarsel-less encodes multiple 9aa polypeptides in a polycistron. I do wonder just how prevalent this type of protein is, as you would’ve thought they’d be found more commonly by classical genetic screens. Anyway could say the same about ncRNAs.
Check out the paper by my former student:
Identification of prokaryotic small proteins using a comparative genomic approach
J Samayoa, FH Yildiz, K Karplus
Bioinformatics 27 (13), 1765-1771 2011