Some omics words we would like to see
Advertisomics: environmental sequencing aimed at obtaining popular press coverage with little or no scientific value. Samples obtained from an environment otherwise not of microbiological interest. “Hey, did you hear they swabbed the car wheels in the building’s parking lot and found that the microbes all cluster by tire brand name?”
Celebromics: sequencing the genome or microbiome of a celebrity. Generally the sequence is not even published, but just the act of sequencing it provides publicity for the lucky lab, the celeb, and maybe even a microbial species or two. “They sequenced the genome of Keith Richards, and found a duplicated set of multiple drug resistance genes.”
Contaminomics: sequencing results published prematurely, and later discovered that the major finding is the result of a contamination.
DuhOmics: unsurprising results from a genomic study. Usually confirming common knowledge that did not require a genomic study in the first place.
Lazarusomics: sequencing the genome of an extinct animal, including hominids, with the implicit or explicit promise that we will be able, very soon, to reverse the extinction.
Shockomics: related to advertisomics. Sequencing for shock value and pop publicity. Usually involving human or animal bodily secretions or parts you’d rather not have known about.
TooMuchInformationOmics: A result of the personal genomics and microbiome industry. No, I am not interested in that heel spur gene that you got from your grandmother, nor am I interested in the novelty of the chlamydia strain they found in your partner’s microbiome.
ZZZomics: an omics paper that makes you fall asleep half way through the introduction.

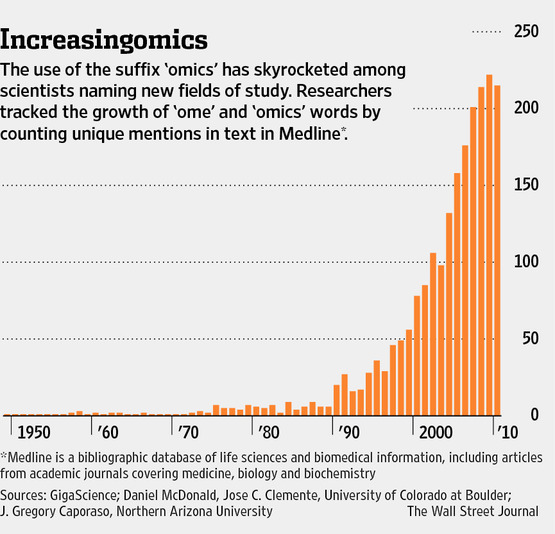


















Don’t forget the “narcissome” or “egome”- researchers characterizing their own ‘omes. Not sure if this tweet will come through in this response- but it’s from Jonathan Eisen.