Can we make accountable research software?
Preamble: this post is inspired by a series of tweets that took place over the past couple of days. I am indebted to Luis Pedro Coelho (@LuisPedroCoelho) and to Robert Buels (@rbuels) for a stimulating, 140-char-at-a-time discussion. Finally, my thanks (and yours, hopefully) to Ben Temperton for initiating the Bioinformatics Testing Consortium.
Science is messing around with things you don’t know. Contrary to what most high school and college textbooks say, the reality of day-to-day science is not a methodical hypothesis -> experiment -> conclusions, rinse, repeat. But it’s a lot messier than that. If there is any kind of process in science (method in madness) it is something like this:
1. What don’t I know that is interesting? E.g. how many teeth does a Piranha fish have.
2. How can I get to know that? That’s where things become messy. First is devising a method to catch a Piranha without losing a limb. So you need to build special equipment. Then you may want more than one fish, because number of teeth may vary between individuals. It may be gender dependent, so there’s a whole subproject of identifying boy Piranha and girl Piranha. It may also be age dependent, so how do you know how old a fish is? Etc. etc.
3. Collect lots of data on gender, age, diet, location, and of course, number of teeth.
4. Try to make sense of it all. So you may find that boy Piranha have 70 teeth, and girls have 80 teeth, but with juveniles this may be reversed, but not always, and it differs between the two rivers you visited. And in River “A” they mostly eat Possum that fall in, but in River B they eat fledgling bats who were too ambitious in their attempt to fly over the river, so there’s a a whole slew of correlations you do not understand… Also, along the way you discover that there is a new species of pacifist, vegetarian Piranha that live off algae and have a special attraction to a species of kelp whose effect is not unlike that of Cannabis on humans. Suddenly, investigating the Piranha stonerious becomes a much more interesting endeavor.
As you may have noticed, my knowledge of Piranha comes mostly from this source, so it may be slightly lacking:
I just used the Hollywood-stereotyped Pirhana to illustrate a point. The point being that I love trashy movies science can be a messy undertaking, and once your start, you rarely know how things are going to turn out. Things that come up along the way may cause you to change tack. Sometimes you discover you are not equipped to do what you want to do. So you make your own equipment, or if unfeasible then look for a different, more realistic goal. You try this, you try that, pushing against the boundaries of your ignorance. Until finally with a lot of hard work and a bit of luck you manage to move a chunk of matter out of the space of ignorance, and into the space of “we probably understand this a bit better now”. This is not to say that science is just a lot of fiddling around until the pieces fall together. It is chipping away at ignorance in a methodical way; in a convincing methodical way: you need to convince your peers and yourself that your discoveries were made using the most rigorous of methods. And that vein of knowledge which you have unearthed after relentless excavation is, in fact, not fool’s gold but the real deal.
Which brings me to research programming.
Like many other labs, my lab looks to answer biological questions that can be answered from large amounts of genomic data. We are interested in how gene clusters evolve. Or how diet affects the interaction between bacteria and the gut in babies. When code is written in my lab, it is mostly hypothesis-testing code. Or mucking-about code. Or “let’s try this” code. We look for one thing in the data. Then at the other. We raise a hypothesis and write code to check it. We want to check it quickly so that, if the hypothesis is wrong, we can quickly eliminate it, but if it appears to be right, we will write more code to investigate the next stage, and the one after that. We slowly unearth the vein of metal, hoping it is gold rather than pyrite. But if it’s pyrite, we want to know it as soon as possible, so we can dig somewhere else. or maybe the vein is not gold, but silver. That would be an interesting side project which would become a main project.
This practices of code writing for day-to-day lab research are therefore completely unlike anything software engineers are taught. In fact, they are actually the opposite in many ways, and may horrify you if you come from a classic software-industry development environment. Research coding is not done with the purpose of being robust, or reusable, or long-lived in development and versioning repositories. Upgrades are not provided and the product, such as it is, is definitely not user-friendly for public consumption. It is usually the code’s writer who is the consumer, or in some cases a few others in the lab. The code is rarely applicable to wide range of problems: it is suited for a specific question asked on a specific data set. Most of it ends up unused after a handful of runs. When we finish a project, we usually end up with a few files filled up with Python code and functions with names like “gene_function_correlation_7” because the first 6 did not work. (I still have 1 through 6 in the file, I rarely delete code since something that was not useful yesterday, might prove to be good tomorrow). It’s mostly throwaway code. that is also why we write in Python, since development time is fast, and there are plenty of libraries to support parsing and manipulating genomic data. More on slice-and-dice scientific coding, why scripting languages are great for it in How Perl Saved the Human Genome Project, penned by Lincoln Stein.
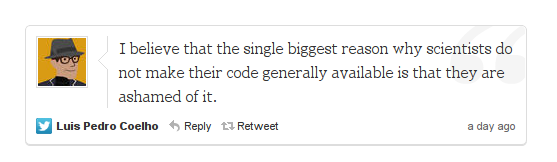
But back to everyday research lab coding. LPC’s tweet that triggered this conversation:
Uncomfortably close to the truth. Not that I am ashamed of my code, it worked great for me! But it would not work for someone else. I’m ashamed to force someone to waste time navigating my scripts’ vagaries.
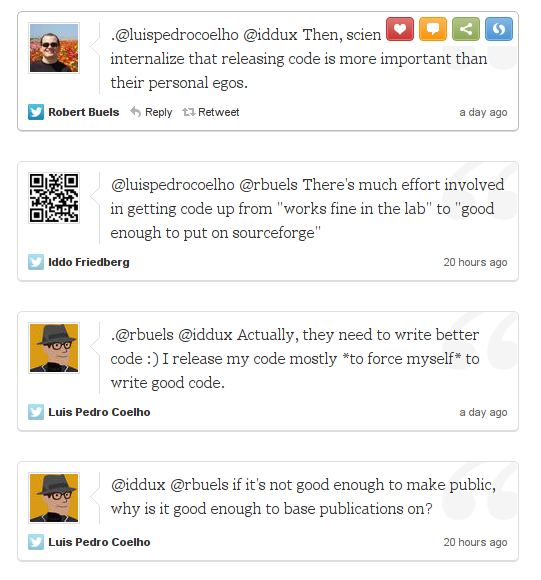
LPC has a point. But again, code which works fine on my workstation can be uninstallable on someone else’s: all those module imports I use, and my Linux is tweaked just so in terms of libraries, etc. Also, I have to write installation & usage documentation, provide module dependencies, provide some form of test input….
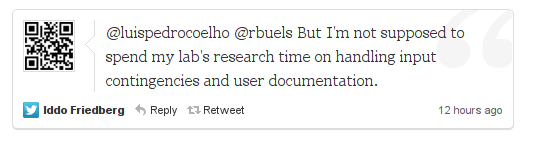
And “by not supposed” I mean “I don’t have the resources”. These things take time, and neither my students nor I have that.
Again, a good point. Can there be some code-verfication standard? Can we distribute our code with the research based upon it without feeling “ashamed” on the one hand, and without spending an onerous amount of time making it fit for public consumption on the other?
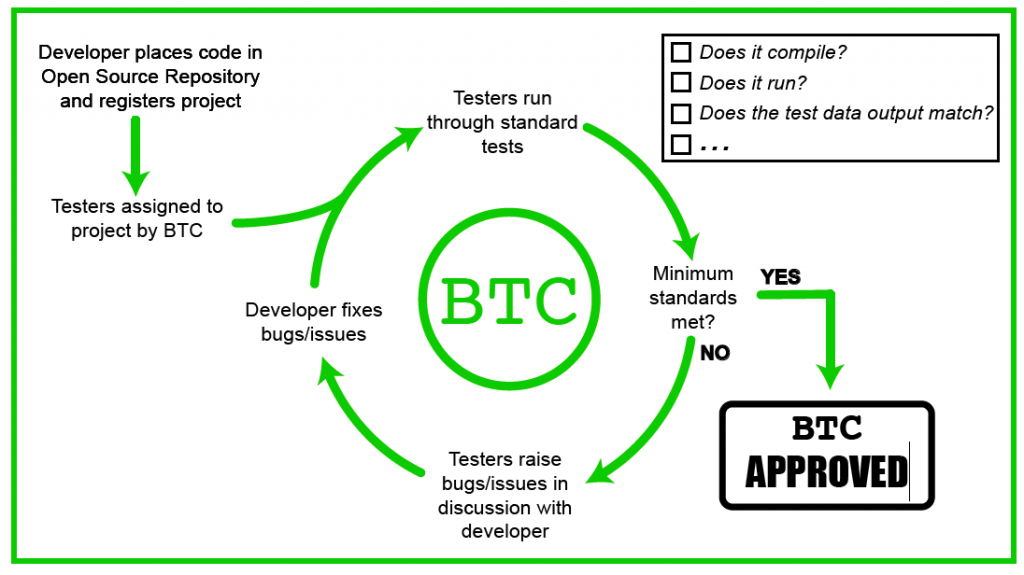
At least for bioinformatic code, Ben Temperton of Oregon State University has come up with an idea: the Bioinformatic Testing Consortium (full disclosure — I am a member):
While the use of professional testing in bioinformatics is undoubtedly out of the budgetary constraints of most projects, there are significant parallels to be drawn with the review process of manuscripts. The ‘Bioinformatics Testing Consortium’, was established to perform the role of testers for bioinformatics software.
The main aims of the consortium would be to verify that:
- The codebase could be installed on a wide range of infrastructures, with identified issues dealt with either in the documentation or the codebase itself.
- Verification of the codebase using a provided dataset, which could then act as a positive control in post-release analysis.
- Accurate documentation of the pipeline, ideally through a wiki system to allow issues to be captured for greater knowledge-sharing.
A great idea, and if taken up by journals, having your BTC-approved code accompanying your paper would go a long way to validating the science presented in your research article. As usual, the problem comes to time and funding: who will be spending them? For now, the suggestion is that testing will be done by volunteers. This may work for a short while, but in the long run funding agencies together with journals should pay some attention to an important lacuna in scientific publishing: the software that was used to generate the actual science is usually missing. If we (publishers, funders, and scientists) are all on the same side, and our goal is to produce quality science, then effort should be made to properly publish software same as effort is being made to publish the results that that software generates. If we pay that much attention to the figures in our papers, we should try to think of way to make transparent, to some extent, the software that made these figures.
Perhaps grant money + a fraction of the publication fees can go towards having your software refined by the BTC and then reviewed along with your manuscript? The thing is, publishing now is quite a laborious process as it is. Preparing acceptable code on top of everything else might push less-resourced labs away from journals that would mandate such practices. Careful thought has to be given as to how research software is made transparent without taxing research labs beyond their already stretched resources.





















Get rid of that spinning tag cloud. It’s incredibly distracting.
I’m afraid I don’t understand your reasoning.
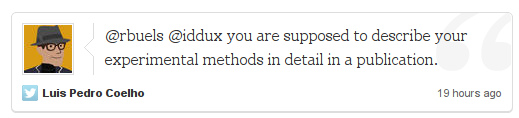
So you “don’t have the resources” (and by resources you mean time) to put your code into some open-source repository. And it takes a few minutes at most to do it (closer to a few seconds, actually, once you have installed the needed tools and registered an account).
And yet, you do have the resources to participate in the consortium that was created to discuss the best ways to make your code available to the community 🙂
I think you worry too much about your code being uninstallable, unreadable, etc. The most uninstallable and unreadable code is the code that is never released. So just go and make it available – maybe you will find some interested users who will help you with testing, documentation and improving quality of your code.
Putting up “raw” code files is really worse than not putting up anything at all. We are talking about a bunch of scripts that are consecutively called through the Python shell, and a large number (most) of those scripts are useless, having historically been relegated to the unworking hypothesis rubbish heap. I would not want to inflict a haystack of code on anyone. Also, some of the code I may not want people to see prematurely. E.g. if that code happens to be in the file, but would be used for a future publication. The idea is to adhere to scientific standards of clear and transparent publication. Putting up this raw code would be akin to putting up a lab notebook, scraps of notes & untested bench protocols, videos of the lab group meetings and snapshots of the lab whiteboard. Scientific communication, at least the “official” kind that goes public, is done through peer-reviewed articles. No one has the time or energy to wade through a lab’s paper- and magnetic- history trail. Plus, few labs will allow it: there is always the next project in the lab’s notebooks and meetings, and no one likes to be scooped.
Cleaning up, documenting, explaining which bit of code was used to generate which results in the paper is very laborious. More laborious than writing the code itself. I am rather surprised that you went through the trouble of reading the post coming out the with impression that I was concerned only about the time it takes to set up a GitHub account. (Which my lab has).
Also, rarely the code stands by itself, and there are usually a few gigabytes of data associated with it. Again, those are messy, not standard, require documentation. So, no, I would not want to put my raw code up there, because no one would get the association to the published research unless they spend an inordinate amount of time and effort doing so. Also, some of it could be harmful to the lab’s competitive ability.
I’m with Alex. It doesn’t have to be perfect, it doesn’t have to run easily on other peoples machines, it doesn’t have to be documented, but it does have to be there. That BTC process looks like a lot of unnecessary red tape. Just because the publication itself goes through a lengthy review process, doesn’t mean the source code has to suffer the same fate. Trust the people who write it to do their jobs properly, trust the community to download it, try it, check it, and critique your work and conclusions.
Frankly, as a software developer also working in scientific research, I have no guarantee your work is not full of mistakes (or worse, entirely fabricated) without access to the code. Given the recent push by Nature to have their results independently verified, in fields where such checking requires labs and great expenditure, complaining about 1 day spent cleaning up the code is laughable.
Oh boy. You’re touching on a point that has been bothering me for a while, both scientifically and personally.
I’m a vanilla computer scientist by training, and have developed a passion for bioinformatics and computational biology after I’ve already spent over a decade working as a software developer and – to make things even worse – an IT security auditor. Since security and reliability are two sides of the same coin, I’ve spent years learning about all the subtle ways software can fail.
Which has given me an unfortunate attention to detail when it comes to my own as well as others’ scientific code. It’s unfortunate because I’m incredibly slow as a bioinformatician. I actually care about code quality, and I can’t bring myself not to care – because otherwise, I have no confidence in the results I’m getting.
During my time working in computational biology/bioinformatics groups, I’ve had a chance to look at some of the code in use there, and boy, can I confirm what you said about being horrified. Poor documentation, software behaving erratically (and silently so!) unless you supply it with exactly the right input, which is of course also poorly documented, memory corruption bugs that will crash the program (sucks if the read mapper you’re using crashes after three days of running, so you have to spend time to somehow identify the bug and do the alignment over, or switch to a different read mapper in the hope of being luckier with that), or a Perl/Python-based toolchain that will crash on this one piece of oddly formatted input, and on and on. Worst of all, I’ve seen bugs that are silent, but corrupt parts of the output data, or lead to invalid results in a non-obvious way.
I was horrified then because I kept thinking “How on earth do people get reliable and reproducible results working like this?” And now I’m not sure whether things somehow work out fine (strength in numbers?) or whether they actually don’t, and nobody really notices.
You wrote:
“It is chipping away at ignorance in a methodical way; in a convincing methodical way: you need to convince your peers and yourself that your discoveries were made using the most rigorous of methods.”
And that is exactly my problem: with the state of things I just described, I’m _not_ convinced that the discoveries were made using the most rigorous of methods. On the other hand, people get the work done, they get results, they get papers past review – so maybe my nitpicking over code quality is misplaced after all?
You state that writing publishable code is a problem of resources, and I very much agree. It’s time-consuming as hell, and not very gratifying, because it’s currently not required _or_ rewarded. But then again, many things in science are a problem of resources. Want to study the, um, say, behaviour of a new type of stoner ciliates under the influence of THC, but your lab doesn’t have the resources to buy a good and suitable microscope, so you decide you’re just going to borrow your grandma’s magnifying glass for this study? You’re probably not going to get away with that.
One of the labs I worked at, the group leader eventually decided to hire a scientific programmer who was then tasked with a) implementing stuff to the specifications of the biologists and bioinformaticians, and b) cleaning up and maintaining code that had previously been written in this group, documenting it, and getting the more broadly useful bits into a releasable state. Sure, that may seem like a luxury, but it turned out to be worth a lot. It freed up the bioinformaticians and biologists to actually do (computational) biology, and it saved time spent on hunting down bugs, or head-scratching over corrupted data.
It’s also noteworthy that having technical assistants in a biology lab is fairly common – which seems to be a matter of the perception of “best practice” in a certain discipline.
At the end of this lengthy rant, I want to add that I like the BTC initiative. Ultimately, we’re talking about developing a “good bioinformatics practice” here, and the BTC initiative may be a step in that direction.
Researchers share tissue samples, case studies, data files, patient records, interview tapes so why not code too?
It doesn’t have to be of commercial quality, just comprehensible. If you can’t be confident that someone else can make sense of your code, how can they be confident in your results?
This doesn’t take much time.
When you write code, write so you can read it in 6 months time.
This is good enough to publish.
[…] Friedberg asks the seemingly reasonable question “Can we make accountable research software?” on his blog Byte Size Biology. As he points out, most research software is built by rapid […]
I’m not sure this is as much of a problem as you make it out to be. When I need to recreate another researcher’s work to do comparisons with my own results, I often just e-mail the researcher or his lab. Unless the research is being conducted on a national defense topic, the code is usually provided right away along with a bit of hand-holding to get it working. I have no problem releasing my code to those who ask either.
The biggest problem in my field (computer vision) is that researchers are patenting their methods, then not providing reasonable means of verifying their results. When the code itself is protected under patent and the researcher or university won’t release it under any circumstances, progress is stifled. Obviously researchers and the institutions who employ them should be able to make a profit on their work, and I don’t know a good solution for this, but it is frustrating to only be provided with a Windows NT, 32-bit dll that can’t be linked in any system I’m using, and can’t be modified to the problem set I’m researching. At that point, I’m forced to rewrite someone else’s code entirely from the publication, and all too often, that isn’t possible due to missing or obfuscated details forced on the publisher by page length restrictions.
I’d like to see more code made available, but at the same time, those who are willing to provide their code do so upon request. Those who are not, won’t under any circumstances, so a consortium or publication requirement isn’t going to fix this problem.
I’m also a computer vision scientist and we’ve recently been looking at the problem of how to publish the experiments that we have run to come up with the results in our published papers. For many years we have been putting together an open source computer vision library (OpenIMAJ) but it’s now expanding into other areas including an experimentation and reference annotation framework.
The idea is that you use the experimentation framework to not only give you all the useful stuff, like cross validation and data set splitting, but the code for the experiment itself is then redistributable such that other people can download it and verify the results. The annotation framework allows citations to be included within the code itself so that as the code runs, all referenced works used within the experiment can be perused. The goal is to have a redistributable encapsulation of an experiment.
As the whole library is written in Java with Maven dependency management, it’s entirely cross platform already.
Obviously, there’s an initial learning curve to using any framework which often puts people off issuing them. They need to provide useful research method implementations (incentive for learning the framework) and be well documented. We’re still in the process of documenting it (as you mention it’s hard getting the resources to spend time doing it rather than research) but maybe this sort of framework should be considered by journals as a recognised and maybe preferred means of code distribution.
[…] sentence quoted above is commentary on a post by a different writer which is summarized as: …most research software is built by rapid prototyping methods, rather […]
[…] on ‘Anecdotal Science’ by CTB » On ‘Research Code’ by Deepak Singh Iddo Friedberg says – Science is messing around with things you don’t know. …. This practices of code writing […]
Where’s the reward in publishing code? At the moment, papers get accepted without code, and referees don’t ask for it.
One of the leading stats journals has a scoring scheme for papers depending on if the code is available, the data is available, and if the editor for reproducibility can run the code on the data and get the published results.
Currently the scoring scheme has no direct input into research impact metrics, but it does encourage people reading the paper to go “ooh, I can just get that code and run the analysis on my data”, and hence are more likely to use and cite it. And getting cited is good for your research metrics…
Nice post. I’m a neuroscientist (and open source developer, as it happens) and I’m currently pushing to encourage code sharing in my field. Not particularly from full-time programmers/computer scientists, but from people collecting and analysing neuroscience data with complex methods. You might find these slides interesting from a talk I gave recently on the topic, mostly focussing on the reasons that people have given to me for why we shouldn’t need to share our code:
https://groups.google.com/forum/#!topic/openscienceframework/o-RaS6AMDMI/discussion
The scoring system that Barry mentions above is the sort of thing that I think we need. Or something following Victoria Stodden’s Reproducible Research Standard (RRS). A little badge of honour for papers that can claim to be ‘verifiable’, by making all necessary code available, should provide just enough incentive to get the ball rolling, I think.
[…] Can we make accountable research software? If the Mars rover finds water, it could be H2 … uh oh! If Curiosity locates H2O, a simmering NASA controversy will boil over. The rover’s drill bits may be tainted with Earth microbes that could survive upon touching water. NY Times discovers that scientists have real lives, too! Vaccination and the 1% (this willful scientific ignorance is a good reason why power shouldn’t be concentrated in the hands of the wealthy) Millipede-eating ant Plectroctena cristata poses with a millipede she has paralyzed Caribbean coral reefs face collapse […]
[…] or technologist thing of getting something working, -ish, good enough “for now”, research-style coding, but it’s always mindful of an engineering style trade-off: that it might not be […]