Now that’s a f***ing big genome!
It isn’t junk DNA: God just commented out a lot of crappy code as he rolled out releases.
— An old bioinformaticians’ joke
(Hey, I never said it was a funny joke…)
Why are some genomes so big? I mean, seriously. Why would the marbled lungfish with a genome weighing 132.83 picograms (pg) need an estimated 130,000,000,000 bp? It may have to do with that fact that these fish undergo metamorphosis, and the large developmental coding this could entail: some amphibians also have big genomes.. then again, some don’t. So the reason for the big lungfish genome is still a mystery.
Then there is the genome of Paris japonica, a rare plant whose genome weighs 152.23 pg, making its genome the largest known so far, at a whopping estimated 150,000,000,000 bp. (Humans have a genome size of 10,000,000,000 3,000,00,000 bp by comparison. Thanks for catching this error, Jason.) Large genomes do not seem to confer an advantage: in fact, plants with large genomes are at greater risk of extinction, are less adapted to living in polluted soils and are less able to tolerate extreme environmental conditions. Their cell-cycle is, of course longer, so they grow slower than plants with a small genome and perhaps also more errors are introduced during mitosis and meiosis. The nucleus size and consequently, the cell size are also bigger, at least in plants. But in their conclusions to the study published in the Botanical Journal of the Linnean Society the authors write that “We are still profoundly ignorant about why some genomes […] are so big and how they operate and function.”
Finally, there are viruses. Not exactly alive, but getting more so as we are discovering viruses with genomes sizes that rival those of bacteria and archaea. I have posted before about the Mimivirus: a virus infecting amoebas which is so large it has been mis-classified as a bacteria for a decade. At 1,181,404 nucleotides its genome may not seem like much compared with Paris japonica and the marble lungfish, but this genome is 100-1000 times larger than that of most known viruses. Mimivirus also has tRNA genes, which are used to assemble proteins and a viral parasite of its own, named Sputnik (“little companion”), all of which makes you wonder whether the working definition for viruses as non-living entities still holds.
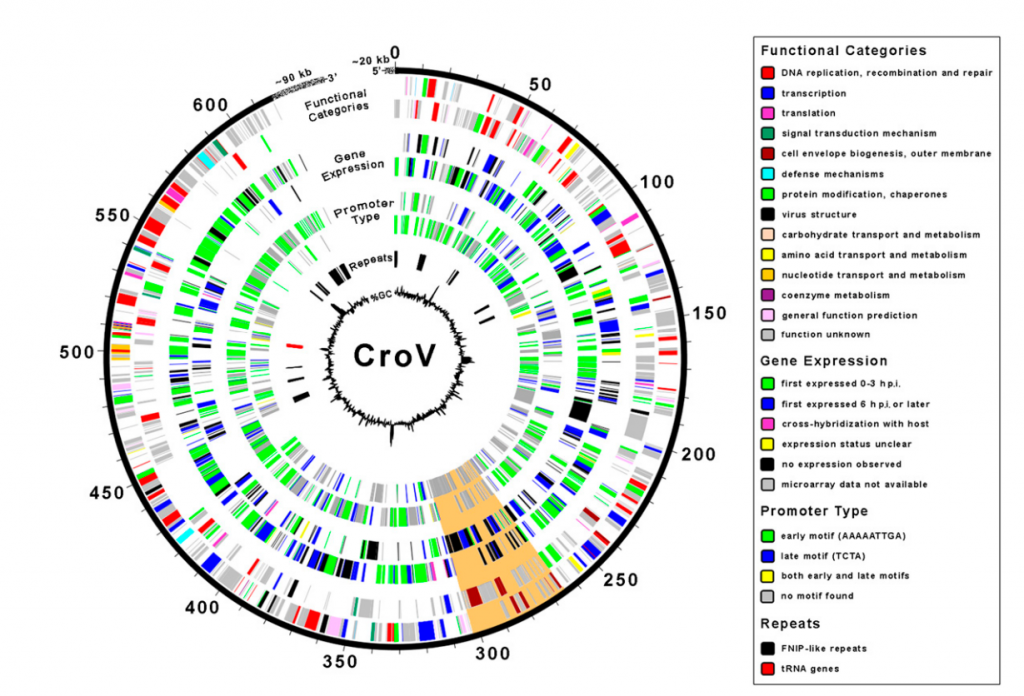
This month, Matthias Fischer and his colleagues have described a large marine ![]() virus, with a genome of 730,000 bp of double stranded DNA. The virus infects a unicellular eukaryotic bacteria eater named Cafeteria roenbergensis. (Why the odd name for the host? “We found a new species of ciliate during a marine field course in Rønberg and named it Cafeteria roenbergensis because of its voracious and indiscriminate appetite after many dinner discussions in the local cafeteria.” Reminds me of Ali G saying that he will name his son after where he was conceived which would be “Langley Village”, with the full name being :’The bogs in KFC in Langley Village’). Hence, the virus infecting this creatively-named critter is the Cafeteria roenbergensis virus or CroV. The virus has some 544 predicted protein coding genes, with at least 274 of them expressed during infection. Among the goodies coded by CroV are transcription related genes, DNA repair genes, promoters, and tRNA. Fairly atypical to known viruses. Which again, begs the question: how much cellular machinery does a virus need to code in its genome to cross the border between life and non-life? Is that even a criterion, or should we also consider the lack of physiology? Still, the majority of genes in CroV, as in Mimivirus and in most known viruses have no similarity to those in “true” living things. Go figure.
virus, with a genome of 730,000 bp of double stranded DNA. The virus infects a unicellular eukaryotic bacteria eater named Cafeteria roenbergensis. (Why the odd name for the host? “We found a new species of ciliate during a marine field course in Rønberg and named it Cafeteria roenbergensis because of its voracious and indiscriminate appetite after many dinner discussions in the local cafeteria.” Reminds me of Ali G saying that he will name his son after where he was conceived which would be “Langley Village”, with the full name being :’The bogs in KFC in Langley Village’). Hence, the virus infecting this creatively-named critter is the Cafeteria roenbergensis virus or CroV. The virus has some 544 predicted protein coding genes, with at least 274 of them expressed during infection. Among the goodies coded by CroV are transcription related genes, DNA repair genes, promoters, and tRNA. Fairly atypical to known viruses. Which again, begs the question: how much cellular machinery does a virus need to code in its genome to cross the border between life and non-life? Is that even a criterion, or should we also consider the lack of physiology? Still, the majority of genes in CroV, as in Mimivirus and in most known viruses have no similarity to those in “true” living things. Go figure.
Fischer, M., Allen, M., Wilson, W., & Suttle, C. (2010). Giant virus with a remarkable complement of genes infects marine zooplankton Proceedings of the National Academy of Sciences DOI: 10.1073/pnas.1007615107
PELLICER, J., FAY, M., & LEITCH, I. (2010). The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society, 164 (1), 10-15 DOI: 10.1111/j.1095-8339.2010.01072.x





















Amoeba dubia and proteus are actually still the largest genomes, at 670GBp for the former (see really nice diagram comparing euk genome sizes here: Keeling & Slamovits 2005 Curr Op Genet Dev: http://www.botany.ubc.ca/keeling/PDF/05COGD.pdf); I fail to find any conclusive reason to mistrust those data, other than the Paris japonica people wanting to lay claim to the largest genome…
Also, the unusually large genome of the lungfish may very well have nothing to do with its metamorphosis; perhaps most genomes are bloated just because they were able to get that way. Lynch & Conery 2003 Science (http://www.indiana.edu/~lynchlab/PDF/Lynch121.pdf) links increase of genome size with decrease of effective population size (and thus, increasing effects of drift relative to selection) in eukaryotes. Perhaps in some cases, a large genome size may be adaptive somewhat, sure. But it seems like the overall tendencies point to it being a result of runaway processes when selection doesn’t particularly mind it too much.
Dinoflagellates are another peculiar group with massive genomes (and not necessarily massive sizes at all – when you look at a dino, there’s this GIANT blob of chromatin staring at you, occupying a significant portion of the cell) – as nobody sequenced a complete dino genome yet, it’s not entirely certain what’s going on, although they do lack most histones. Additionally, EST projects reveal that there’s a lot of junk accumulating from their peculiar splice leader trans-splicing mechanism and reverse transcription… really bizarre stuff. I must say, plants and animals are fucking boring as far as genomes go! =P
Happy to see viruses brought up though – loads of interesting stuff lurking amidst non-medically important viruses, who deserve way more attention than they get!
@psi_wavefunction The authors of the Japonica genome article actually mention the Amoeba dubia . They claim the measurement may be vastly inaccurate since it was not estimated using current molecular methods. See also http://www.science.smith.edu/departments/Biology/lkatz/documents/McGrath_Katz_Tree.pdf
I agree with this. links increase of genome size with decrease of effective population size (and thus, increasing effects of drift relative to selection) in eukaryotes. Perhaps in some cases, a large genome size may be adaptive somewhat, sure. But it seems like the overall tendencies point to it being a result of runaway processes when selection doesn’t particularly mind it too much.
Hmmm… this has become a Keeling vs Katz issue now – both are great experts on genomes + genome size, and both are good scientists, so I wonder whom to believe in this case. Exacerbated further by major conflict of interest issues… >_>
It’s hard to distinguish polyploidy from large genome size without sequencing the genome, I guess. Furthermore, after several whole genome duplications… yeah, I can see it getting messy. Speaking of which, how did the plant people distinguish polyploidy from genome size? Or are we ignoring that altogether? Because if so, then ciliate macronuclei and radiolarian primary nuclei are pretty intense in size. As are foram nuclei, to some extent. The protist world is orders of magnitude bigger than any other eukaryotic kingdom, so I’d strongly suggest rigorously investigating the genome sizes there before claiming the largest genome among plants…