I can’t hear you, the bacteria are too noisy
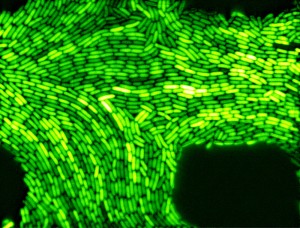
Much too noisy. When looking at a population of genetically identical bacteria, the number of proteins they produce varies. The picture below shows the levels of one type of protein that was fused to a green fluorescent protein (so we can see it): clearly there is a variation in how much of the protein each cell produces (“protein expression” in molbio-speak), even though the bacteria are genetically identical. Why is that? In 2006, a group of researchers at the University of California San Diego and Boston University looked at the variation in protein expression in genetically identical bacteria, and what it could mean. They constructed a simple and well-defined computational model first. The researchers were surprised when their model shows that the variation actually increased when the cell growth and division was slowed or stopped. This prediction seemed paradoxical: if the cells are less active, how come the variation in protein expression increases? Shouldn’t they all be going into some “baseline production mode”? To answer these questions, they took them to the lab. Nicholas Guido and his colleagues engineered bacteria with simple gene networks, where the production of the gene could be induced, repressed and both induced and repressed simultaneously. The gene itself was with green fluorescent protein, so that the more protein is produced, the brighter the cells shine under light. Lo and behold, the computational predictions were correct! (1) the expression of the protein was not uniform (even though the cells were genetically identical) and (2) variation in protein expression increased when the proliferation of the bacterial cells was slowing down or has stopped.
Their explanation to this random noise in protein production: the need for variation to survive. Bacteria often deal with quickly changing conditions: temperature, oxygen concentration, water, chemicals, antibiotics… all these can kill. If the population is identical, what kills one kills all. But if even within a genetically identical population there are variations in protein level expression, then the population is not phenotypically identical even though it is genetically so. Some bacteria may survive the dry spell, the heat or — what concerns us quite a bit — the onslaught by antibiotics. The random population variation or “noise” in protein level expression is an evolutionary survival mechanism.
Fast forward from 2006 to last week. In a brilliant work published in Science, Yuichi Taniguchi and colleagues from Harvard University and University of Toronto looked at individual E. coli cells for protein expression. They used examined different strains, 96 at a time using a microfluidic chip. Each lane on the chip has room for a single cell, enabling them to quantify the levels of proteins in single cells from the same or different strains very quickly. Taniguchi and colleagues examined 1018 different genes in E. coli which covers about 25% of the genome. Like Guido and colleagues, Taniguchi and colleagues also found a large variation in the expression of the same protein in different cells which were otherwise genetically identical, no matter what the protein was. They also found that different kinds of proteins were produced in different distributions in the cell. They also measured was noise: how much randomness went into the production of proteins. What they found were two kinds of noise: one type of noise was from proteins that were produced in small numbers (less than 10 molecules per cell) the more protein produced, the lower the variation in protein production, or “noise”, between cells. A second type was from proteins that were produced in larger numbers. For those, there is a “noise floor”: the fluctuation in protein production does not decrease below a certain point, and there is less fluctuation in proteins that are produced in high numbers than in those produced in low numbers. This means that the cellular mechanisms of protein degradation and/or production control may hit some sort of steady-state once protein production reaches a certain level.
They did not stop by looking at proteins, though. In each cell, they also looked at the level of mRNA coding for that protein. mRNA production numbers are also very noisy: actually, noisier than those of protein. But surprisingly, Taniguchi discovered that when looking at single cells, mRNA and protein levels do not correlate. Not even a weak correlation, and no matter what the protein. The high noise levels and lack of correlation in expression can be explained by the different lifetimes of protein vs. mRNA. mRNA is quickly degraded in the cell, while proteins may outlive cell-division. mRNA is produced in short bursts, “lives fast and dies fast”, while buffering protein levels from high fluctuation levels.
Looking at these studies together, we can say that there is a lot of noise in the system, but it serves a purpose: not only on the selection level (as discovered in 2006), but also on the systems level (as shown last week). On the selection level, noise fosters differences between individuals, which gives at least someone from the bacterial population a chance to survive if conditions change drastically. It is less clear what is happening in the level of the intracellular system: for proteins expressed in large numbers, it seems like there is some external control mechanism at work that keeps noise above a certain level.
How does mRNA and protein production noise then propagate, say, across gene expression pathways, when one protein can cascade the production of many others? How much is noise a control mechanism on a cellular and cellular-ensemble level? Are there “noise clamping” and “noise amplification” mechanisms that need yet be discovered? The Taniguchi study hints that there are, and the Guido study strongly suggests that they are affected by the environment. I think we are only beginning to hear the noise bacteria make.
Taniguchi, Y., Choi, P., Li, G., Chen, H., Babu, M., Hearn, J., Emili, A., & Xie, X. (2010). Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells Science, 329 (5991), 533-538 DOI: 10.1126/science.1188308
Guido, N., Wang, X., Adalsteinsson, D., McMillen, D., Hasty, J., Cantor, C., Elston, T., & Collins, J. (2006). A bottom-up approach to gene regulation Nature, 439 (7078), 856-860 DOI: 10.1038/nature04473




















If protein expression levels and mRNA levels are really uncorrelated, as you seem to be implying, it throws into question all the microarray and RNA-seq data. My understanding was that they are correlated, though far from identical, due mainly to different rates of degradation for different proteins.
Incidentally, you may be interested in checking out my blog and adding it to your list of science blogs.
I have a very similar picture of my son:
http://users.soe.ucsc.edu/~karplus/abe/oct-nov-96/Abe-headphone-head.jpeg
@gasstationwithoutpumps
Well, the Taniguchi paper clearly states that they are uncorrelated at the single cell level. Here is a link to the relevant figure in the paper:
http://www.sciencemag.org/cgi/content/full/329/5991/533/F4
You’re right, it does run counter to many things we know and assume. Also, I am not sure if single-cell transcription /translation noise & protein / mRNA correlation has been checked on tissue data (as opposed to bacteria in this study). Tissue data is where many RNASeq and quantitative proteome data comes from.
Oh, and I didn’t know you had a blog Kevin… sorry about that. It’s great, I’ll add it to my blogroll.
@gasstationwithoutpumps
Protein and mRNA levels are often uncorrelated. Some of the first systems to be investigated were the AOE (anti-oxidant enzymes) in mice ( http://www.ncbi.nlm.nih.gov/pubmed/8639268 ). Recent studies on RNA localisation has really underlined that mRNA is really not a good indication of protein level. Then, of course there is the issue of “functional” protein. Many proteins are present, but not activated.
Hand me that microarray and pass the salt.
One of the most fascinating pieces of work to come out last month! Maybe we will look back in a decade or so and be amazed that we ever thought mRNA levels were a good proxy for protein expression..
Coincidentally, this is also on of the clearest blogposts I read on the subject. Since I’m hosting next edition – would you consider submitting it to the Molbio Carnival (http://blogcarnival.com/bc/cprof_10473.html)?
@Lucas
Seems like the captcha in the blog carnival submission site is on the blink… so I cannot submit. You are welcome to submit it yourself, of course. And thanks!