Filling in the evolutionary blanks, genome by genome
After hearing Jonathan Eisen and Nikos Kyripdes talk about GEBA in various meetings, it is great to see the paper finally come out, and under a CC license too. Good move for everyone.
GEBA is the Genomic Encyclopedia of Bacteria and Archaea. The idea is simple: we have >1000 prokaryotic genomes in GenBank as of today. But those were sequenced under a myriad of interests: clinical, functional, ease, biotechnological or pharmaceutical potential, etc. In evolutionary terms, those 1000 genomes provide a very biased view of the tree of microbial life. That would be like sampling mammalian life in Europe and North America only: you would miss out on most big cats, Elephants, Rhinos, not to mention all the marsupials. To correct this situation, teams from the Joint Genome Institute, UC Davis and several others set out to perform a more uniform sampling across the tree of prokaryotic life. The first batch of 56 genomes from GEBA is published today in Nature; fifty-three bacterial and three archaeal.
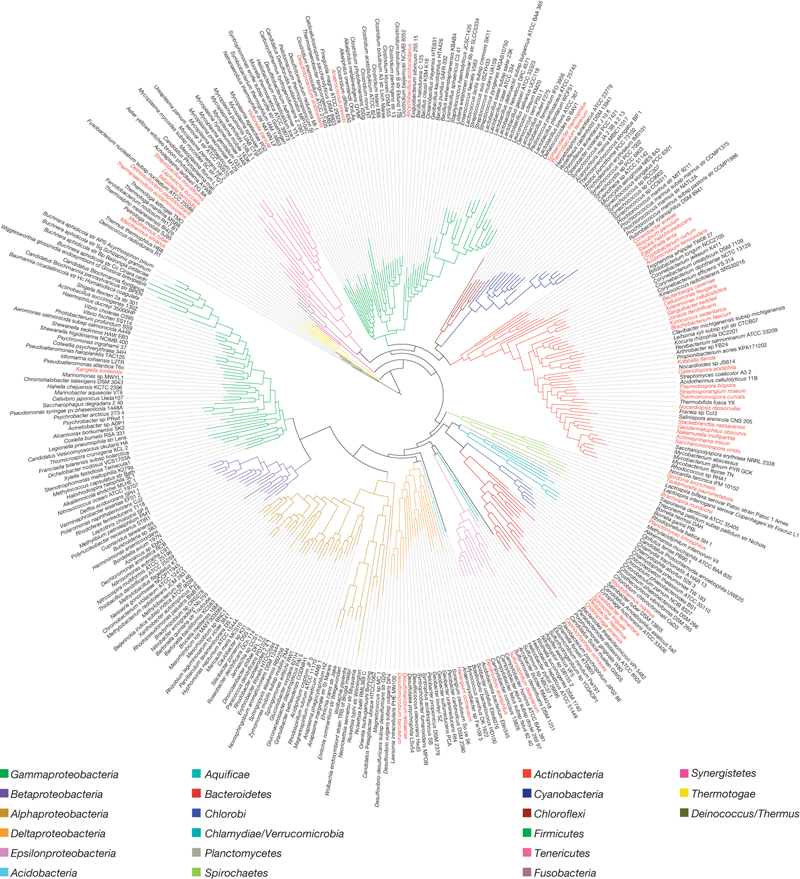
Maximum-likelihood phylogenetic tree of the bacterial domain based on a concatenated alignment of 31 broadly conserved protein-coding genes. Phyla are distinguished by colour of the branch and GEBA genomes are indicated in red in the outer circle of species names. Click to open original in Nature.
It seems that they are on the right track to enrich our understanding of bacterial genes and genomes using this phylogenetically-mindful sampling strategy. For example, they show that their sampling enables the discovery of an average of 1,060 protein families/genome. Sampling a single bacterial family would provide 121 new protein families, sampling within a bacterial phylum would give an average of 308 new protein families, and within a bacterial domain, 650. They have discovered a total of 1,798 families that seem to have no similarity to any existing family, hinting at new bacterial functionality (or maybe some new prophages?) They have discovered a few new cellulases, genes that break down cellulose, the polymer that makes up plant cell walls. Cellulases are the holy grail of the biofuel prospecting industry: specifically, a cellulase that can be exploited en-masse to turn plant matter into fuel economically. They also discovered a homolog of Actin, a cytoskeletal protein thought until now to only exist in eukaryotes.
One thing that is sorely missing is accessibility. Yes, the individual genome papers are all published in SIGS and in Nature under open access, which is great. But when you go to the GEBA site, you get a simple description of the candidate genomes. The annotations are somewhere behind a password-protected site, but I could not seem to get an account to view them. A proper genomic browser for the sequenced and annotated genomes, with some phylogenetic map showing who is located where on the tree would go a long way towards helping the rest of us explore this new comprehensive picture of prokaryotic genome space.
Finally, if you want to hear more about how they did what, here’s Eisen talking about GEBA.
Wu, D., Hugenholtz, P., Mavromatis, K., Pukall, R., Dalin, E., Ivanova, N., Kunin, V., Goodwin, L., Wu, M., Tindall, B., Hooper, S., Pati, A., Lykidis, A., Spring, S., Anderson, I., D’haeseleer, P., Zemla, A., Singer, M., Lapidus, A., Nolan, M., Copeland, A., Han, C., Chen, F., Cheng, J., Lucas, S., Kerfeld, C., Lang, E., Gronow, S., Chain, P., Bruce, D., Rubin, E., Kyrpides, N., Klenk, H., & Eisen, J. (2009). A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea Nature, 462 (7276), 1056-1060 DOI: 10.1038/nature08656



















Working on the JGI-GEBA pages – not sure why it is password protected from where you went. But I note, the data is all in Genbank. And it is also all in Biotorrents at http://www.biotorrents.net/details.php?id=38.
Also – the better GEBA site is http://img.jgi.doe.gov/cgi-bin/geba/main.cgi
Try that and you should find the genomes (I hope …)
Thanks Jonathan.
Both kinda lead to the same place. The GenBank (and Biotorrents) presence is well noted. But since GEBA is all about exploring genome space, I was half-expecting a clickable version of that tree you have in Figure 1, + some comparative genome browsing? JGI has the infrastructure for that with the IMG.
Well, the genome space browser is a great idea — but would be some work — I will suggest it to IMG people and to others — might make a good grant proposal —
I note, the numerical gotcha thing has issues when I click “back” after I forget to type it in once …
IMG does have some annotation browser. Linking to that seems easy. Granted (pun intended), making a more sophisticated tree-able browser is somewhat non-trivial…
The numerical gotcha is the least suckiest antispam mechanism I could find. All the graphical captchas take time to load, which makes commenting a drag…
I was going to test out my new research blogging account by reviewing this paper. Oh well, I guess some may think I am a little biased being in his lab.
Nice review, and I think there is still lots of improvements to me made for viewing and analyzing comparative genomic projects.
Morgan -you can’t be as biased as I am and I just posted a “research blogging” article about this paper too … still figuring out research blogging’s system though so not sure it is working right