Photosynthesis, phages and structures: there’s treasure everywhere!
Here’s a really cool work, published this September in Nature.. Why did I choose this work? Well, it’s a major discovery, and it’s all done using bioinformatics, and fairly simple bioinformatics at that. The power of metagenomics and bioinfromatics: in a mass of data you just have to know what you are looking for, and how to look for it.
Viruses as a bacterial genetic mechanism
Viruses follow some interesting and sometimes convoluted evolutionary paths. One is “infect quick, reproduce fast, and make sure you can get to the next host before you kill this one”. That is pretty extreme: smallpox was doing that, when there was smallpox. Ebola is doing that, but not very well: killing the host too quickly means that the disease is contained, especially in rural areas. Another strategy is: “slow and easy wins the race”. The herpes virus does that. Not lethal, but laying dormant in the central nervous system, it is infectious, but rarely causes anything more than they occasional cold sore (which admittedly, is painful and disturbing). Still, it manages to infect up to 90% of the human population, most of which are completely unaware they harbor it, and would never develop any symptoms.
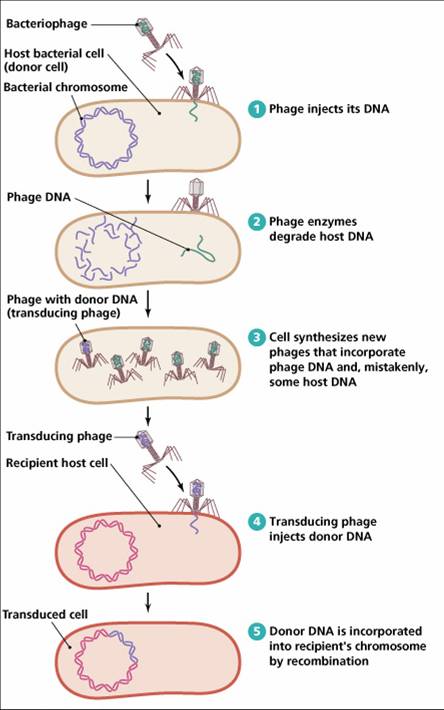
Most of the viruses on earth don’t infect humans, nor animals, nor plants. They infect microbes, where the same spectrum of evolutionary strategies applies. Some attack quickly, killing the microbial population they infect. Other can remain dormant for a long time. It is becoming clear to us that bacterial viruses or bacteriophages, are responsible for a large portion, if not the majority, of genetic variance in bacteria. In fact, viruses are a major component in bacterial genetics. The mechanism is called transduction, and it is illustrated below. Bacteriophages pick up DNA from bacteria they infect, and transfer it to other bacteria, creating genetic variance in the bacterial population.
Viral transduction also adapts
But viral transduction does not just carry random genes. Natural selection favors transduced genes that increase the bacterial host’s fitness. Because when a bacteria is infected by a virus, its protein making machinery is used to make viral genes. But when the viral genes include genes that are beneficial to the host as well, then everybody wins: the phage-infected bacterial species gets genes which enable it to compete better for resources with other bacterial species, while the phage gets a larger number of hosts to infect. Of course, this has to go hand in hand with a relatively benign virus that remains dormant long enough to let the bacterial host species enjoy the benefits of the transduced genes.
Such is the case of cyanophages and cyanobacteria. Cyanobacteria are photosynthetic bacteria, and cyanophages are the viruses that infect them. Several studies have shown that cyanophages have acquired whole photosynthetic genes from bacteria. Viruses do not photosynthesize, but when they infect cyanobaceria, the viral photosynthetic system is added to the bacterial one, boosting bacterial photosynthetic activity and ultimately increasing bacterial energy production.
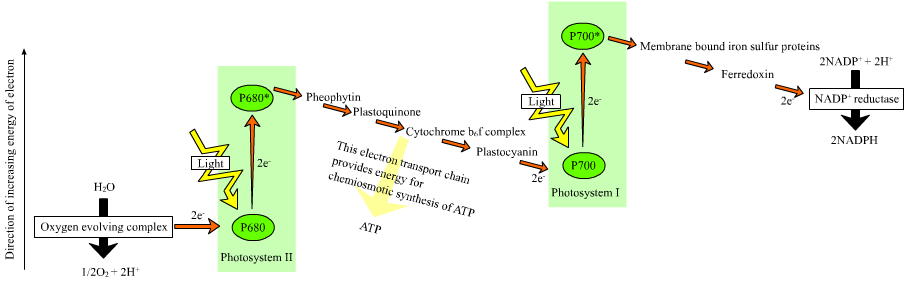
The photosynthetic mechanism is divided into two components: photosystem I and photosystem II (PSI and PSII). For a few years now, PSII has been known to be transduced by cyanophages.
A more recent study by Itai Sharon and colleagues published in Nature this September shows that PSI proteins are also tranduced by cyanophages. Also, it seems like the viral PSI has some interesting properties that may make it advantageous over the cyanobacterial PSI. Two proteins in the bacterial PSI are called PsaJ and PsaF. They found that the homologous protein in cyanophages is a fusion of the two, PsaJF. When they modeled an insert of PsaJF into the bacterial photosystem I it seemed that the bacterial PSI with the viral insert can now function more efficiently than the the original bacterial PSI. As a rule, PSI is a system that accepts electrons from PSII via a protein called plastocyanin. The donated electrons are excited by light, and the energized electrons are used to synthesize ATP and NADPH, the energy coinage of the cell, which are used to synthesize sugar from CO2. However, when the bacterial PsaJ and PsaF are replaced by the viral compound PsaJF, it seems like plastocyanin does not have to be the only electron donor to the newly minted virally-donated PSI. This means that the PSI may now accept electrons not only from plastocyanin, but from other electron-carrying proteins as well. Such proteins that are involved in the respiratory system, for example, which also donate electrons. The advantage of such a setup is that electrons whose reducing power would otherwise go to waste, got through PSII for formation of extra NADPH and ATP. Sharon and colleagues do not prove all this experimentally, but they make a pretty strong case, citing some analogous cases.

a, The structure of T. elongatus PSI (subunits) was illustrated by PyMOL (http://pymol.sourceforge.net/) using a PSI monomer (adopted from Protein Data Bank (PDB) accession 1jb0). PsaF is in magenta, PsaJ is in blue, and all of the other subunits are in green. b, A model for the structure of the viral PsaJF fusion protein (red) substituting the original PsaF and PsaJ subunits. Reproduced under NPG Licensing terms for non-commercial / educational purposes. doi:10.1038/nature08284
Like I said, this work is purely bioinformatics. They basically mined the Global Ocean Survey metagenomic data, over six million sequences from marine microbes collected by the J. Craig Venter Institute which I mentioned in another post. They then identified sequences that contain PSI genes, and sifted through those to find sequences that also contain genes that are exclusively viral. Having both a PSI gene and a viral gene on the same DNA clone ensures they were taken from a virus. I’m not sure how they did the structural modeling and insertion of the PsaJF. This seems to be missing both from the Nature article, and the supplementary material. Yes, it’s one of those Nature works with 3 pages of article, and 28 of supplementary. Great read though, there’s treasure everywhere.
Sharon, I., Alperovitch, A., Rohwer, F., Haynes, M., Glaser, F., Atamna-Ismaeel, N., Pinter, R., Partensky, F., Koonin, E., Wolf, Y., Nelson, N., & Béjà, O. (2009). Photosystem I gene cassettes are present in marine virus genomes Nature, 461 (7261), 258-262 DOI: 10.1038/nature08284
Lindell, D., Jaffe, J., Johnson, Z., Church, G., & Chisholm, S. (2005). Photosynthesis genes in marine viruses yield proteins during host infection Nature, 438 (7064), 86-89 DOI: 10.1038/nature04111
Comments are closed.