Using phylogenetics to reconstruct a 59 million year old drug
Good news:
Press Release
2011-10-03
The Nobel Assembly at Karolinska Institutet has today decided that
The Nobel Prize in Physiology or Medicine 2011
shall be divided, with one half jointly to
Bruce A. Beutler and Jules A. Hoffmann
for their discoveries concerning the activation of innate immunity
and the other half to
Ralph M. Steinman
for his discovery of the dendritic cell and its role in adaptive immunity
(Unfortunately, Steinman died between the committee’s decision and the announcement. He still received the Prize, though.)
However, it is not news (and not good) that we are losing the arms race against bacteria. We are overusing antibiotics in medicine and in agriculture, virtually nurturing today’s and tomorrow’s killers. A report in Wired earlier this year paints a bleak picture:
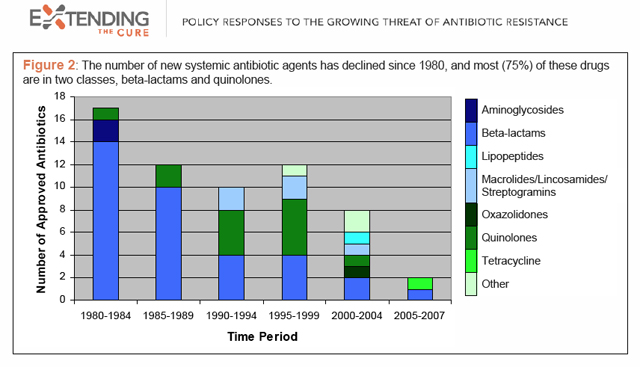
Truly new antibiotics are critically needed because bacteria, having no experience of them, cannot immediately mount resistance to them — something that does happen with me-too compounds featuring some slight molecular change. But they’re rare. As this chart from the research group Extending the Cure shows, antibiotic development has slowed dramatically over the past 30 years, and among the few drugs being brought forth, most share the mechanisms of already-existing classes.
To understand the extent of the crisis, we have to remember that antibiotics are the foundation of a huge number of medical procedures, from cancer treatment to dentistry. Taking away this foundation would cripple modern medicine. Together with vaccines and public hygiene, antibiotics are the reason that many of us live longer — and better — than our grandparents’ generation and before that.
So lacking drug company motivation (more on that in McKenna’s report) and facing a dwindling supply of effective antibiotics, what are we to do?
A good start is to take a second look at nature. For example, here is a Kangaroo being born:
Awww…
The interesting thing about Kangaroos and other marsupials is that their young are being exposed at a very precarious stage to the outside world. Roo is born in a fetal state: underdeveloped, blind, barely moving and has little to no body temperature regulation. All of these problems are taken care of by Kanga’s protecting Roo’s six minute trip to her pouch where things are warm and cozy, and maternal milk flows in abundance. All except one: pathogens. During his trip, Roo can acquire a whole bunch of nasty bugs from Kanga’s fur and the air. Also, the pouch is not exactly a sterile environment and there is a possibility of infection there. So how is little Roo to survive the trip to the pouch and subsequent stay?
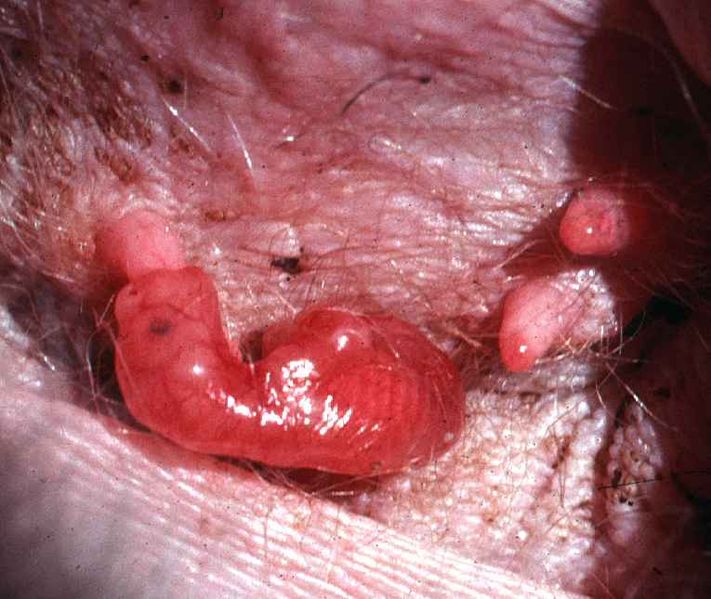
Joey in pouch. Photograph by Geoff Shaw (Zoology, University of Melbourne, Australia). From Wikimedia Commons.
The answer is innate immunity, that collection of mechanisms which protect the host in a non-specific manner. While his adaptive immunity is quite undeveloped, Roo does produce some killer all-purpose peptides he can use against microbes. The same class of peptides are produced in Kanga’s milk. Collectively they are known as cathelicidins. Only about 30 amino acids long, these highly charged molecules kill both gram-positive and gram-negative bacteria.
Since the tammar wallaby genome has recently been sequenced, a group of Australian scientists has decided to hunt for cathelicidins in the tammar’s genome. They also looked for cathelicidins in the genomes of the duck-billed platypus (a monotreme) in the opossum (an American marsupial), in human, mouse, cow & sheep (all placentals). Here is what they found. Each leaf on the tree represents a cathelicidin gene. The leaf colors and shapes are for different species of origin.
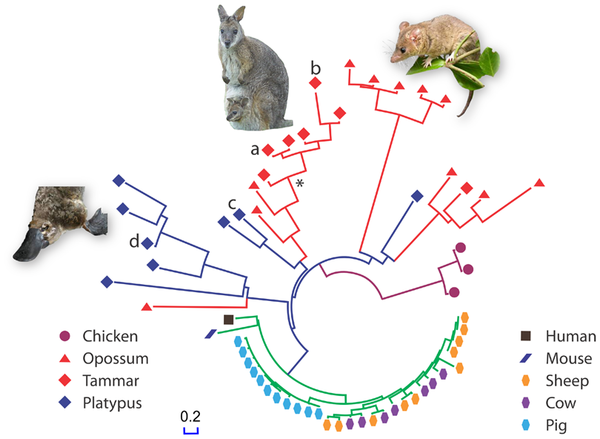
From Wang J, et al. 2011 PLoS ONE 6(8): e24030. doi:10.1371/journal.pone.0024030 Reproduced under CC license.
The marsupials and monotremes have a much higher diversity of cathelicidin genes than the placental mammals. Note that there are many duplicates of the cathelicidin gene in Pig and Cow, but these duplications are very recent as we can tell by the highly similar sequences of the genes. These duplications are probably because herd animals are more susceptible to pathogens. In any case, pig and cow have higher copy number of cathelicidin genes, and thus perhaps a large number of proteins, but not many different ones as in marsupials and monotremes. Diverse cathelicidin genes in the genome can offer a wider protective umbrella against many different types of pathogens.
Back to the antibiotic crisis: can we use marsupial cathelicidins as an antibiotic in humans? Do cathelicidins kill human pathogens? Are they toxic to humans? The authors checked the first two. They used cathelicidins from wallaby and platypus to kill human pathogens: P. aeruginosa, K. pneumoniae and A. baumanii, including antibiotic resistant strains. Cathelicidins were much more effective than, well, antibiotics against those bacteria. Also, cathelicidins did not kill human red blood cells, which makes them a potential drug. Of course, immune reaction against cathelicidins as a foreign still needs to be checked, among many, many other things, but the whole idea of looking at marsupials is that, as mamals, they may be able to supply us with clues on how to synthesize a cathelicidin to be used as a drug in humans.
![]()
The really cool thing about this study is that the authors used phylogenetics to design an ancestral wallaby cathelicidin. They aligned all the wallaby cathelicidin protein sequences, which were conserved in 40 of the 46 amino-acid positions. They used PAML, GASP and Ancescon to reconstruct an ancestral wallaby sequence, estimated to be 59 million years old, marked by an asterisk (*) in the above gene tree. They then synthesized the ancestral WAM (Wallaby Anti Microbial) peptide and used it to kill bacteria. Guess what: the ancestral WAM was even better than the existing WAMs, as even a lower concentration was needed to kill bacteria. They are still working on red blood cell toxicity (yay for PLoS-enabled comments!)
So maybe the answer to the increase in bacterial resistance to antibiotics lies in Kanga’s pouch. After all, she was very protective of Roo.
EDIT: clarified that the part about synthesizing and testing ancestral-WAM to kill bacteria.
Wang, J., Wong, E., Whitley, J., Li, J., Stringer, J., Short, K., Renfree, M., Belov, K., & Cocks, B. (2011). Ancient Antimicrobial Peptides Kill Antibiotic-Resistant Pathogens: Australian Mammals Provide New Options PLoS ONE, 6 (8) DOI: 10.1371/journal.pone.0024030



















Thanks for attracting my attention to this paper, really cool. I wouldn’t say that it’s a 59 MY old drug, it’s actually much older. We could say that kangaroos have 59 MY of experience fine tuning it, though.
Project idea: reconstruct and test the ancestral version against bacteria.
Hi Marc. Actually, they did that, from the post: “They then used the ancestral WAM (Wallaby Anti Microbial) peptide to kill bacteria. Guess what: the ancestral WAM was even better than the existing WAMs, as even a lower concentration was needed to kill bacteria. They are still working on red blood cell toxicity (yay for PLoS-enabled comments!)”
I edited the post a bit to make this clearer.
We wanted to try to help them with this by reconstructing the indel history too, but failed because the highly-variable terminal exon of the cathelicidins defeated our indel reconstruction methods at the time. I think now we have slightly better methods and would do a better job. Very glad to see this work even though I am a bit sad we couldn’t help with it.
Thanks, this is really cool.