It ain’t necessarily so
First, a short glossary.
Homologous genes are descended from a common ancestral gene.
There are two types of homology:
- Orthology is homology due to a speciation event. So if there is a gene A’ in humans and A” in mice, and they are obviously similar in sequence, we infer that they homologous. We usually also infer that they are orthologous, as the common gene ancestor A existed in the common ancestor of humans and mice, some 600 million years ago. Once the ancestral lines diverged, the genes carried over into the respective progeny.
- Paralogy is homology due to a duplication event. A gene has been duplicated in a species genome, and the genome now has two copies of this gene in place of one.
Orthology, Paralogy and Function
It has been proposed that paralogous genes would generally have different functions. The rationale being that in-species duplication, two copies of the same gene are redundant. One copy maintains its function, while the other is “free to explore” other functions. The flipside of this hypothesis is that orthologs maintain functional similarity, because the progeny species inheriting the orthologous genes need to maintain their function.
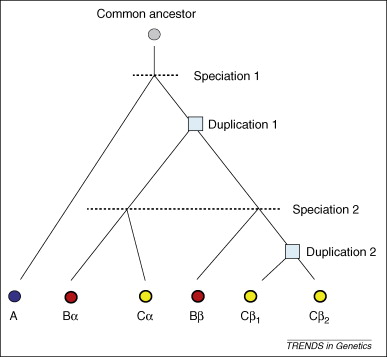
Formation of orthologs and paralogs. The evolutionary tree shows six homologous genes from three species designated A, B and C. Genes are represented by circles and each color represents a different species; genes with paralogs are circled by a thicker line (only the gene in the A lineage does not have a paralog). Boxes at nodes represent duplication events. Duplication 1 produced paralogs α and β in the ancestor of B and C, whereas duplication 2 produced paralogs β1 and β2 in the C lineage. All genes from B and C are co-orthologs to the gene from A. Genes α and β are in-paralogs relative to speciation 1, but are out-paralogs relative to speciation 2. Genes β1 and β2 are in-paralogs relative to both speciations in the tree. Genes Bα and Cα are one-to-one orthologs. From doi:10.1016/j.tig.2009.03.004
Functional innovation through duplication has been hailed as a major driving force in evolution. After all, it is hard to accept the Darwinian tenet that random changes — even if directionally selected — can constantly produce innovative complexities. A duplicate gene provides an already existing complexity. Imagine many such duplications, and you can see how duplicate genes provide an genomic “functional opportunity bank” for the biosphere.
Only, maybe not. Romain A. Studer and Marc Robinson-Rechavi challenge common wisdom by publishing a study that says: “it ain’t necessarily so”. They look at three alternative models of molecular function evolution: (i) subfunctionalization after duplication; (ii) neofunctionalization after duplication; and (iii) the ‘alternative model’ of equal change after duplication or speciation. Subfunctionalization holds that after duplication, each of the two copies of the gene performs only a subset of the functions of the ancestral single copy. Neofunctionalization holds that one of the two genes possesses a new, selectively beneficial function that was absent in the population before the duplication. The ‘alternative model’ states that the gain of new function is not preferential to paralogs and that orthologs may gain new functions at the same rate that paralogs do.
Studer and Robinson-Rechavi claim that few studies have been made to study the scope of any of these proposed models. They then lay out study designs for doing so, challenging other evolutionary biologists (and themselves?) to conduct these studies and examine whether the common wisdom that orthologs maintain function while paralogs gain function. What I like about this paper is that it not only makes a strong case for challenging conventional wisdom, it also lays out a series of possible routes of study to be taken up by others.
Update: MK pointed out an obvious lacuna in this post:
Studer, R., & Robinson-Rechavi, M. (2009). How confident can we be that orthologs are similar, but paralogs differ? Trends in Genetics, 25 (5), 210-216 DOI: 10.1016/j.tig.2009.03.004
Comments are closed.


















