The new natural history
Before the 20th century biology was, to a large extent, “Natural History”. It was an observational rather than the experimental science it is considered to be today. At that time, the typical biologist, a natural historian, was going about the (European colonized) world, collecting specimens of new and fossilized species, classifying and recording them for posterity. Armed with small pick-axes and hammers for extracting fossils, ether-laced jars for insect specimens, formaldehyde for preserving tissues, butterfly nets, drawing pads in lieu of cameras, guns and various other paraphernalia. In some cases he (no “she” in science at the time) went to field carrying his own hypothesis about nature and her laws and looking to prove it. But in mostly it was the gathering knowledge for knowledge’s sake. Reported in natural societies’ meetings and compiled in tomes of encyclopedias. Every marine exploration voyage carried its own naturalists, as well as many merchant and naval ships. From the plethora of data, the patterns and laws describing life emerged. The most famous of the theories developed by a naturalist was evolution by natural selection. Darwin went through years of observation and classification while developing his landmark theory.
Towards the end of the 19th century, biology began to be studied on a molecular level. Chemists have become interested in the underlying molecular machinery of life, and the field of biochemistry was born. Armed with a keen experimental philosophy of science, and the tools to execute it, they took biology into the lab. Decomposing the basic processes of life, and the molecules that they facilitate and act upon. Formulating hypotheses, testing them with controls, executing rigorous protocols to discover natural processes of life. Biology has also become intertwined with medicine through microbiology and physiology. Again, experimentation came to the forefront, as medicine adopted scientific means to study diseases and develop treatments. Within the realm of larger scales of life science, ecology has in many ways inherited natural history, transformed into a science of understanding the interactions among living things.
And then came molecular biology. With the discovery of the structure of DNA and the genetic code, many of the mechanism by which life is encoded and perpetuates its information have been deciphered. We consequently learned how to manipulate genetic material at the level of a single nucleotide or amino acids, studying the function of genes by studying artificial mutants; expressing genes of one organism in another. Molecular biology has enabled to study life at a basic level, creating carefully controlled environments not dreamed of by the natural historians whose scientific mandate was to observe and record. Biology has reached an experimental apex. We also learned how to easily and cheaply read, or sequence, genes.
Today we are sequencing whole genomes on a regular basis. The genome of the first free living organism, H. influenzae was sequenced in 1995. Human in 2001. Rat, mouse, nematode, fruit fly, water cress and other model organisms had their genomic sequences read, with the hope of understanding what they do. The plethora of information brought about the field of bioinformatics: computational biology applied to analyzing informational biomolecules, such as nucleic acids and proteins. Sequencing technologies became exponentially cheaper over the past 50 years. Genomes and metagenomes are now sequenced as a routine matter. A typical prokaryotic microbe can be sequenced for under $1,000. Whole microbial communities can now be “fed into” sequencers, to extract the genomic information not of one, but of thousands of microbes in one sequencing run. A century after the dusk of natural history, biologists are again going outside to collect knowledge for knowledge’s sake. This time the natural historian is armed with microbial and tissue collection kits and freezers in the field, and sequencing machines and cluster computers in the lab. She (yay progress!) is going to alkaline lakes, mine shaft drains, termite guts and cow rumen to sample the microbial life. Indeed,the soil and water of the Galapagos, whose flora and fauna inspired Darwin have recently been revisited by another ship. This time, the microbes, not the Finches, were studied. Back in the lab, the specimens are not mounted, Rather, their DNA, RNA and proteins are sequenced. Terabytes of information are stored, while methods better storage, retrieval and analysis of the growing volume of data are developed. Universities and research institutes have sequencing centers, and a whole industry of sequencing service centers is flourishing and growing.
Biology is descriptive yet again. Biologists are recording data for posterity. They are cataloging. Natural history is back. The fraction of papers reporting new genomes is constantly growing. Indeed, many new genomes are not even reported in papers, but rather deposited directly into genomic databases. Encyclopedias are back in fashion. Only this time around, they are not large volumes of books taking up shelves in the library of the scholar’s home or university office. Rather, they are web-sites, holding more raw data than whole university libraries, unconstrained by the cost of paper and printing labor, and usually free for anyone to explore: the genome databases. Genomic encyclopedias exist for all large genomes, and for the smaller ones that are found interesting. The other genomes that are generated on a daily basis are incorporated into the larger all-encompassing such as GenBank which contains virtually all the genetic and genomic data that has been made public.
There are differences between the natural historians of yore (is 100 years “yore”?) and the sequencing natural historians of today. Chiefly, the nature of the data has changed: from a purely informatic point of view, we are not dealing with hundreds or even thousands of new animal and plant species. We are dealing with billions of bytes of character data associated with thousands of animals and plant species, and an uncounted number of microbial species. The information is extracted from this data by complex and constantly evolving computational means. The following questions are typically asked: what is the actual genomic sequence? (Sequence assembly). Where are the genes? (Gene finding). What is there function? (Functional annotation). How are they activated? (Computational systems biology on the one hand, and structural biology on the other). What is the genotypic variation in the community and how does this variation contribute to evolution and adaptation of a population? (Metagenomic / metadata analysis). Each one of these questions has spawned a discipline that is concerned with optimizing its ability to extract the relevant information (e.g. gene location, gene function) from the raw genomic data.
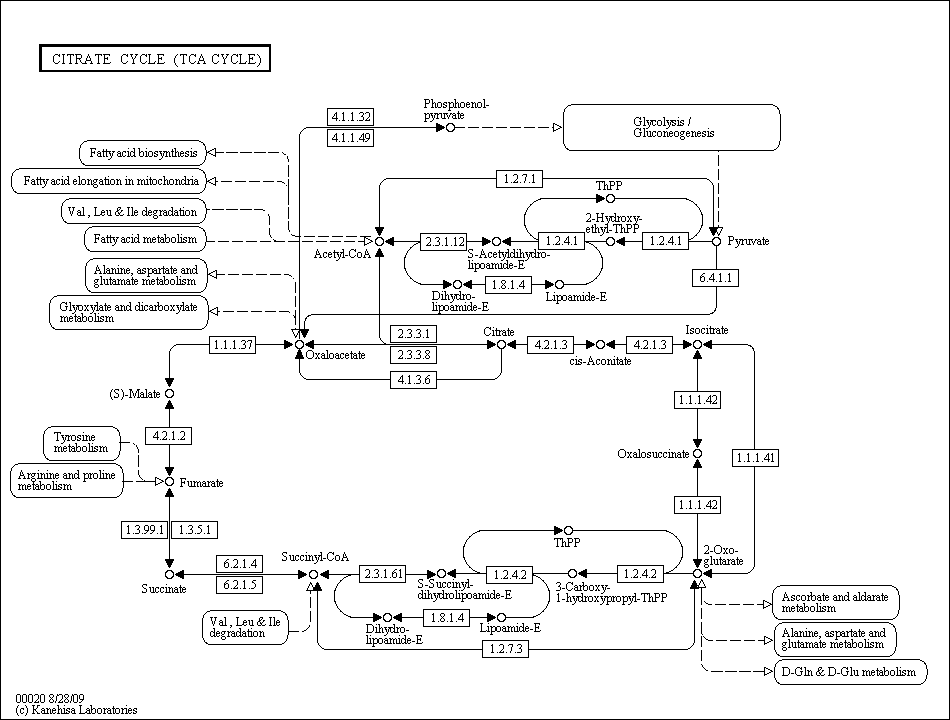
The TCA cycle from the Kyoto Encyclopedia of Genes and Genomes. Click for the full encyclopedia entry.
Welcome back, natural history! Observing and describing the richness and diversity of life has always been part of biology. With genomics and metagenomics, descriptive biology, after being somewhat back stage for so long, is back in the limelight.
Comments are closed.