A FLORA of Protein Structure to Protein Function
Proteins are the machinery of life, and they facilitate most of life’s functions. Traffic into and out of the cell? Protein pumps, pores and channels. Respiration? Proteins. Metabolism and catabolism? Proteins. Immune system, signaling, development… all complex networks of interacting proteins. Understanding a protein’s structure can tell us a lot about how it performs its function. If we know what a protein does, we can look at it’s molecular workings, and generally figure out how it does it. Hemoglobin carries oxygen in most animals, something that has been known since 1840. However, it is only when Max Perutz and John Kendrew solved the structure, that the actual mechanism of oxygen binding and release has been elucidated. Since Perutz’s and Kendrew’s discovery in 1949, the structures of some 35,000 proteins have been solved.
When we know the protein’s structure we know a lot about how it performs its function.That would be the equivalent of looking at a diagram of a car engine, and then exclaiming: “oh, so that’s how it works!” But the converse does not hold true. If we have the structure, we may not be able to infer the protein’s function. Imagine having the diagram of a new engine which you have never seen before. It might be a car engine, but which make and model? Or it might not be a car engine at all, but that of a lawnmower, or a boat, an electric generator. The point is, without knowing what the diagram represents, we would only have a general idea that we have a machine that burns some sort of fuel to power something.
We face the same problem with protein structures. It does happen that we solve the structure of a protein, whose function is unknown. Oh. Kay. What now? We are stuck with a diagram for a machine which we do not know what it does. Therefore, any kind of method we can devise to predict a protein’s function from its structure would be very helpful. Christine Orengo’s group at University College London, UK has been tackling this problem for quite a while. Her group has recently published a paper in PLoS Computational Biology where they describe an algorithm that can classify engines enzymes: a subgroup of proteins that catalyze chemical reactions. The classification algorithm works as follows:
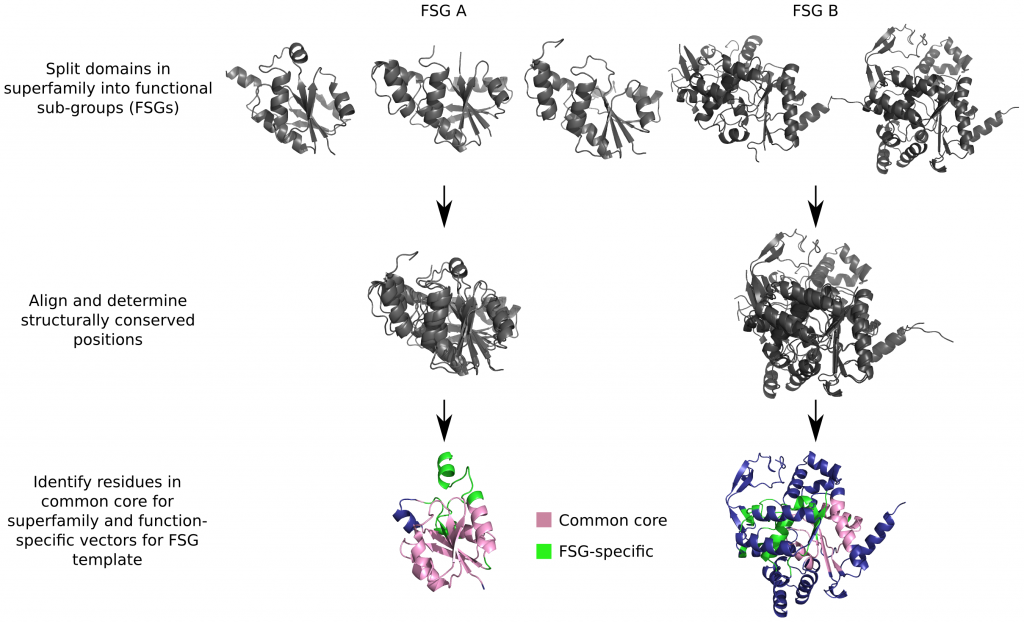
1) They partitioned all enzymes of known function into functional subgroups, or FSGs. Within an FSG, all proteins have the same function. Two proteins from different FSGs will have different functions.
2) Next, they selected a set of conserved vectors from a given domain in a given FSG which, when compared against relatives of different functions/FSGs, would produce a low score. Conversely, when proteins from the same FSG are compared, they should have a significantly higher score.The vectors are measurements of distance and direction along the side chains of conserved amino acid residues. They found that this differentiating set of vectors is best obtained when the proteins are aligned within and between FSGs, and the vectors are taken from the conserved residues in the FSG alignments.
3) Once they determined which vectors are more conserved within a given functional sub-group (FSG), they created a library of conserved vectors within FSG, a sort of an FSG bar-code. Although the constriction is technically unsupervised, limiting the vectors to conserved residues within an FSG naturally lands them with lots of active site residues.
Having created the template library, they can now find vectors on test proteins, and scan those against the library of conserved vectors, using a simple similarity function. Although (or because) their method is quite simple, they receive very high sensitivity and precision. The methods they compare against are all global structure aligners (such as CE and CATHEDRAL), and by virtue of simply adding spatial information of the conserved / functional residues they greatly improve the function annotation. The great thing about this work is the jump in improvement by adding this very simple, yet so far mostly neglected, attribute.
Unfortunately, no software yet. Too bad because…..
Redfern, O., Dessailly, B., Dallman, T., Sillitoe, I., & Orengo, C. (2009). FLORA: A Novel Method to Predict Protein Function from Structure in Diverse Superfamilies PLoS Computational Biology, 5 (8) DOI: 10.1371/journal.pcbi.1000485





















Re. availability…
We have a FLORA web service planned — hopefully aiming at around the end of this year or the beginning of 2010. We figured this would be an easier way to make it available than a software distribution. As is often the case in bioinformatics, it’s a loose collection of tools, scripts and datasets rather than an easily distributable package. The web service will be free and unrestricted (unless anyone misuses it!) like all our services.
Feel free to contact me for any more information via http://biotext.org.uk/ — although I didn’t work on the algorithm I will be working on the service side of things.
Here is a database that allows a user to search for certain structural motifs of active sites:
http://www.ebi.ac.uk/thornton-srv/databases/CSA/
I realize that this doesn’t predict the entire function of a protein, but could be used as an interesting stepping stone.