Tardy Reading and Scientific Breakthroughs
A friend of mine who is also a scientist once told me: “the only time I read, is when I write”. What he meant was, that the only time he reads scientific literature, is when he writes his own papers, and needs to do the proper research to place his research in the context of the general happenings in his field. Of course that is an exaggeration, as he wouldn’t be as successful as he is without keeping up-to-date. Nevertheless there is something in what he says: in our line of work there is a strong tendency to gloss over or skip literature that is not of immediate interest. We do most of our “scholarly reading” when we write, or prepare a seminar or a class. I read quite a bit when refereeing manuscript: not just the manuscript I received, but also several related papers, so I can assess the novelty of what I am reading. So refereeing is actually my chance to keep abreast of my field, and when treating it as such, it becomes less of a chore.
That being said, I finally got around to reading the December 18, 2008 Science issue that includes last year’s breakthroughs, according to Science‘s editors, with the intent to blog about what I find there. Then I thought, “OK, if I am always late at my reading, how about I blog not about last year’s breakthroughs, but about those of a decade ago”? It would be interesting to look what was hot 10 years ago and see if and how things have changed.
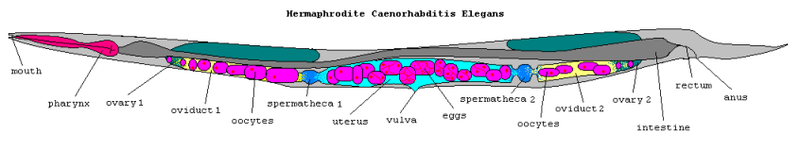
I am a biologist, so I will only treat the breakthroughs that are somewhat related to my field I can understand. 1998 was the year the draft genome of C. elegans was completed: the first genome of a multicellular organism. Canenorhabditis elegans is a nematode, the size of a grain of rice, and an incredibly popular model organism. It is easy, cheap and fast to breed, can be frozen alive and thawed for later use, cell lineages have been documented so they are great for differentiation studies. There are more reasons having to do with this worm’s simplicity and tractability. The number of identified genes was reported to be 19,000, with about 6,000 whose functions were unknown. A quick look today reveals that 1034 current genes are categorized as “Unknown” (go to the “Current only” tab). The total number of predicted genes is 21188.* So some pretty good progress was made in finding out what those genes do. Huzzah. Also, C. elegans is still going strong as a model organism: a simple Google Scholar search reveals that a whopping 10% of life-science articles published in the years 1998-2008 have “C. elegans” in their title. In the years 1997-1987 only 4% had the same. So C. elegans is definitely not going away soon.
Some microbial genomes were completed that year, including T. pallidim (syphillis), Chlamydia trachomatis (causes trachoma and vaginal infections) and Mycobacterium tuberculosis (tuberculosis). Ten years down the line, has this helped us combat those diseases better? In the case of Syphilis this is almost a moot question since T. pallidum is one of the few pathogenic bacteria that still responds to penicillin. The spread of Chlamydia and TB and access to treatment has to do a lot with social and economic causes; more so than medical ones.
At the same time, understanding the genomes of pathogenic bacteria carries a benefit that is broader than finding better treatments those specific diseases. Each genome added to the pool increases our understanding of the mechanisms of pathogenicity, its evolution, and how different microbes interact with their hosts. In the arms race that is going on between us and drug resistant microbes, genomes are key for staying one step ahead. The more genomes we have, the more context we have; we just keep in filling in the evolutionary blanks. Also, more drug targets can be found. Targets that are species-specific, avoiding the need for broad spectrum treatment which accelerates the appearance of resistant strains. We now have the sequences of over 500 microbial genomes, and we have moved to metagenomics, an expansion of genomics that enables us to study genomic profiles of microbial populations and communities.
DNA microarrays made it big in 1998. Microarrays, or DNA chips are used for several types of genomic analyses. Each array contains thousands of microscopic spots that can hybridize with DNA or reverse-transcribed RNA from a sample; they provide a raw picture of the analyzed genome. DNA microarrays can be used for single nucleotide polymorphism (SNP) analysis, transcript expression level analysis, and resequencing mutant genomes.
As a first year PhD candidate in that year, I remember many of my peers going into DNA microarrays. The field had great appeal, especially if you came from a computer science background: simple clustering and graph analysis algorithms that have been used in computer science for years could be easily applied to data from DNA chips, with some spectacular results. Today, microarrays are still going strong, although next-generation sequencing seems to be taking over some of the applications, especially resequencing and SNP profiling.
Lab-on-chip, or microfluidic-based diagnostic devices were also hailed as a breakthrough, and lumped together with DNA chips. A bit odd, as aside from some miniaturization aspects, those technologies are rather different. Also, while DNA chips enjoyed a huge success in the past ten years, the lab-on-chip had a more modest success.

Potassium channel: The protein is displayed as a green cartoon diagram. In addition backbone carbonyl groups and threonine sidechain protein atoms (oxygen = red, carbon = green) are displayed. Finally potassium ions (occupying the S2 and S4 sites) and the oxygen atoms of water molecules (S1 and S3) are depicted as purple and red spheres respectively. License: image is in public domain; text reproduced under CC license; click on image for Wikipedia entry.
OK, there were other interesting things. The first structural solution of the structure of a potassium channel that revealed the mechanism for its specificity; Roderick MacKinnon and Peter Agre shared the Nobel prize for this discovery in 2003. They answered the following question: a potassium ion is larger than a sodium ion; so how does a potassium channel let through the larger potassium ions, but not the smaller sodium ions? The answer lies in the charge density of sodium, that creates a larger water shell around it. The potassium ion is effectively smaller when going through a charged channel, since the water shell around it is smaller.
The further elucidation of the circadian cycle control genes CLOCK , TIM PER and DOUBLETIME took place in 1998, although in my humble and uninformed opinion, the main breakthrough was achieved a year before, when CLOCK itself was discovered. Then again, circadian rhythm research was very much in vogue in the 1990s. My take on this is that scientists are people who travel a lot and suffer from chronic jet lag: fixing circadian rhythms then becomes a top priority. Not that there is a cure for jet lag yet.
The number one breakthrough, the breakthrough of the year 1998, was in astrophysics, and literally dwarfs the breakthroughs made in biology, important as they may have been, and still are. That was the discovery that the universe was expanding at an accelerating rate, making us wonder where the excess energy to support this expansion is coming from, and setting the stage for (Added later: I would like to clarify that when I said “literally dwarfs” I was not talking of discovery importance, whatever that means, but of sheer size: the universe is bigger than C. elegans).
Well then,that is all fine and dandy, but what about 2008’s breakthroughs? Well, too bad I only read when I write, and I really have to stop writing now. Maybe in the next post. Ta-ta.
Comments are closed.