Making genomes less CAGI
cag·ey /ˈkājē/ (adjective)
Reluctant to give information owing to caution or suspicion
CAGI /ˈkājē/ (acronym)
Critical Assessment of Genomic Interpretations. For details keep reading.
The ability to sequence one’s genome adds a new dimension to the ancient maxim “know thyself”. What could be more revealing of one’s self than one’s own blueprint, explaining existing traits such as flat feet or pollen allergies, and problems to come such as male baldness, aging diseases or cancer risks. Yet, a decade after the sequencing of the first human genome, and despite identifying thousands of variants which are associated with different human conditions ranging from earwax constitution to heart disease, we have gained very little in our ability to predict or treat diseases using information in an individual’s genome. Also, although our genetoic makeup plays a large role in our response to drugs, it is rarely considered when dosing drugs. (Warfarin being one of a few notable exceptions.) Genetic testing for known single-gene based disorders using genetic markers has been around for almost two decades, and that is about the extent of our ability to test and interpret genomic data. Yes, we have resources such as SNPedia, but for many genetic variants the association with a phenotype, if any, is very weak.
One major cause for the gap between our genome and, well ourselves, is that we cannot yet interpret the genome to use that information. We do not even know to what extent that information can be used to diagnose and cure diseases. The genome has often been compared to a book. Well, inasmuch as that analogy is somewhat flawed, what we have right now is a book in a foreign language. We know (in many cases) where the words start and stop: those are the genes. We understand some words. But after this our knowledge peters out. Think of idioms, allusions puns or double-entendres that are the unique product of a language, and may be misinterpreted by a non-native speaker.
Freedonia’s Secretary of Treasury: Sir, you try my patience
Rufus T. Firefly: Don’t mind if I do. You must come over some time and try mine.— Marx Brothers “Duck Soup”
The words “try” and “patience” have one meaning in the Secretary’s sentence, and another in Rufus’s sentence. The humor in the exchange would be lost on anyone who does not understand the two context-dependent meanings of those words. It may also be lost if you do not like Groucho Marx’s style of snappy comebacks.
A similar thing happens when we try to understand the function of genes. We may know that a certain gene is an enzyme, but how does this one enzyme affect us? And how do different mutations in the enzyme affect us? Certain mutations may not affect function at all. Others may affect, but only under certain environmental conditions, or in concert with other genomic variants, or depending upon the type of cells in which this enzyme exists. Can we predict the effects of different mutations computationally? In other words, can we interpret the genome?
Enter CAGI, or Critical Assessment of Genome Interpretation. Steven Brenner and Susanna Repo from the University of California, Berkeley and John Moult from the University of Maryland organized a competition between groups to see how well bioinformaticians can predict the connection between a genotype and a phenotype. From the website:
The Critical Assessment of Genome Interpretation (CAGI, \’kā-jē\) is a community experiment to objectively assess computational methods for predicting the phenotypic impacts of genomic variation. In this experiment… participants will be provided genetic variants and will make predictions of resulting molecular, cellular, or organismal phenotype. These predictions will be evaluated against experimental characterizations, and independent assessors will perform the evaluations. Community workshops will be held to disseminate results, assess our collective ability to make accurate and meaningful phenotypic predictions, and better understand progress in the field. From this experiment, we expect to identify bottlenecks in genome interpretation, inform critical areas of future research, and connect researchers from diverse disciplines whose expertise is essential to methods for genome interpretation.
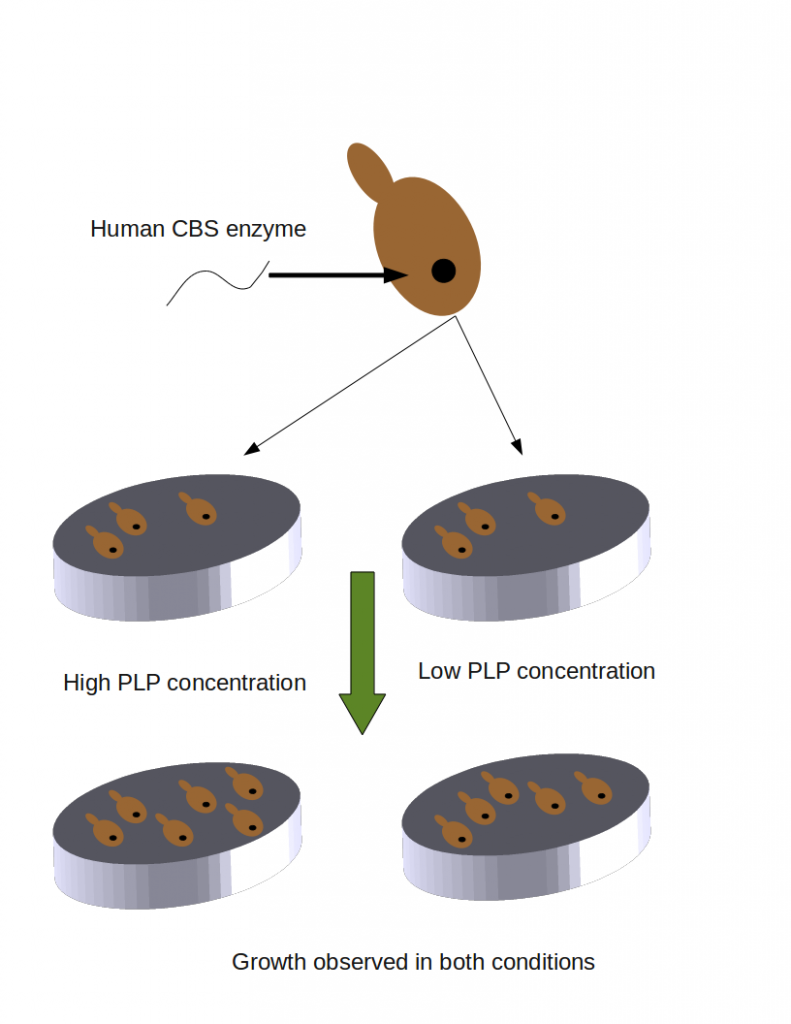
Over the course of three months, Brenner, Moult and Repo received experimental results of mutations from assays intended to examine the functions of several different genes. They revealed some of those results so that bioinformaticians performing computational predictions could train their methods. But most of the experimental results were hidden, to be revealed only at the CAGI meeting itself, some time after the predictors submitted their predictions. Besides predictors and experimental data providers there were also assessors: people who received both predictions and the experimental results, and scored the predictions. I was tasked with being an assessor for the predictions made for one of the data sets. The assayed gene was cystathione beta-synthase or CBS. CBS is involved in the synthesis of the amino acid cysteine, and needs to bind vitamin B6 as a co-factor to function properly. Jasper Rine and Dago Dimster-Denk from Berkeley placed different mutants of the human CBS genes in yeast, and examined their growth in media containing low or high concentrations of PLP, a precursor of vitamin B6. In some cases the yeast grew well in high PLP concentrations but not in low ones. In some cases they did not grow at all, and in some cases the yeast grew just fine. Their were many in-between cases in which yeast growth was less than 100%. The predictors tried to to predict how any given mutation would affect yeast growth; more details of the CBS yeast growth assay are here. You can also read about the other assays involving p53 (a tumor suppressor gene) CHEK2 (associated with breast cancer), and others. Pauline Ng from the Genome Institute of Singapore and I were tasked with scoring the predictions of the CBS assay. We had less than two weeks in which to come up with methods, code them up and implement them. Lots of work, (many thanks to Gaurav Pandey for his heroic and invaluable contribution the night before CAGI!) but lots of fun: we had full creative leeway, and came up with some interesting solutions as to how to score these the 21 methods from about 15 groups who submitted their predictions. Unfortunately I cannot go into the details of our methods here, as they are the subject of an upcoming publication. But I will revisit this topic after the paper is published.

Growth of yeast cells with one type of mutant: no growth in low PLP concentrations. Predictors were asked to predict the growth rate of yeast cells transformed with 50 different mutants and grown in high and low PLP concentrations.
The CAGI meeting at Berkeley was fun, with lively debates springing up about the different prediction methods, assessment methods, where and when the next CAGI meeting should be held. Brenner and Repo were very gracious hosts, providing ample food, drink and downtime to lubricate the scientific discourse. AAA+++ will definitely go again.
One last thing: the best predictors received CAGI Molly as a prize. If there was one point on which everyone agreed it was that CAGI Molly should continue to be the prize. See for yourselves why:
Thanks to Yana Bromberg for the vid.
UPDATE: see also Nature News. Dancing Molly & all.
Comments are closed.