My own post genomic moment
![]()
Maybe I am slow on the uptake, but I never quite liked the term “post genomic”, and I used it very sparingly. (Yes, I do have that term in one of my better cited papers, smack in the first sentence of the abstract, but I never liked that). Perhaps because of all the associated abuse and hype that vacated the term from any core meaning it may have originally held; or perhaps it was because I saw genomics as an ongoing endeavor that will be embedded in life science for a long time to come, with no obvious “post” planned. I even view metagenomics — hailed by many as a completely new and exciting field — as an extension to genomics, with no clear boundary separating the two disciplines. So for me, a bioinformatician, genomics started somewhere around the mid 1990s when whole genome sequences started coming out, it is ongoing, and will continue, no “post-” in site.
Until now.
For I have seen the light, I had an epiphany, a revelation, and I will spread the word. I will shout “post genomics” from the mountaintops; I shall proclaim the word to all I see; for I am a convert, and a true believer in post-genomics.
As with many a conversion, the trigger was seemingly modest. No huge brilliant Nobel-laureate essay in Nature; no national academy symposium (although I have been to one, it was excellent, even though they kept using the “P.G.” word all the time); no road-to-Damascus haloed appearance of J. Craig Venter wielding a green laser pointer showing me the True Path. My own post-genomic moment was triggered by a software article in BMC Bioinformatics. Warren and Setubal from the Virginia Bioinformatics Institute at Virgina Tech published a bacterial genome annotation pipeline article that has taught me the true meaning of “post-genomics”. Yea, verily, I say unto you.
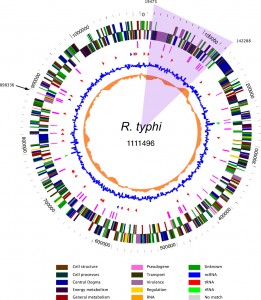
What is a genomic annotation pipeline? Once you sequence the genome of your favorite organism, and assemble it into a whole chromosome or chromosomes, you have the actual template for understanding that critter staring you in the face. Nevertheless, all you really have is a very long string composed of four characters: A, G, C and T. There are no immediately obvious pointers as to where the information is. Where does a gene start? Where does it end? And once you find a region encoding a gene, you still have to find out what it actually does. In broad brush strokes, genome annotation consists of two major steps:
- Structural annotation: find the location of genes and other functional elements on the sequence.
- Functional annotation: assign biological functions to those elements.
Every genome project invests a major fraction of its time and money in annotation. It often uses several pipelines; those can be home-coded, taken from others, and usually a mixture of both. Genome annotation is a major science industry in the (now “post-“?) genome era. This Wikipedia entry is a good place to start learning more on this topic.
So what is so special about a genomic annotation pipeline? Aren’t there quite a few around, genomics being as big as it is? Yes there are but they are mostly inaccessible to the actual end users. They are usually well embedded in the machines of the major model organism genome projects. Or they are made downloadable, but installation is laborious, manual intervention is necessary in many cases, and the process is generally kludgy and heavily human-embedded. This is actually good if you are carefully annotating a new genome, especially a complex animal or plant one for the whole world to see. You are actually somehow a part of a group of curators, bioinformaticians, annotators and scientists on the job.
But what if you have sequenced (or had someone else sequence) your own rather simple bacteria as part of your research into this bug, and you would like to annotate it yourself? You are not a bioinformatician, you do not have the resources to construct or even install and operate a heavy, manually intensive pipeline. Nevertheless, you would like to see a good first draft of an annotated genome: you will probably focus on the interesting spots later, but you would like a rough map of the genomic terrain first. That is where Warren and Setubal’s Genome Reverse Compiler comes in handy: they developed a minimum-hassle genome annotator. I downloaded and installed it on my laptop in about 5 minutes; a simple collection of Perl scripts and C++ code, batteries included. No esoteric CPAN libraries to download, no reliance on previous packages you cannot get from your own Linux distribution, and probably already have. (Ubuntu 8.10 in my case). The architecture is modular: there are several easily understandable components, that can all be lumped together into a single pipeline, or used separately one at a time, or even as part of other pipelines if you are so inclined.
The annotation pipeline relies heavily on heuristics: both for structure annotation and for function annotation. This means that if you have a good evolutionary spread of related organisms already annotated, GRC intelligently copies those annotations over to your bug, with some standard caveats. I like it that in the functional annotation part GRC takes care not to necessarily use the highest BLAST hit for functional transfer, but uses a GO terms (if given) to refine the decision
So the use of GRC can be as manual or as automated as you like. That being said, there are still a few easy steps the writers can take to really streamline the process of installing and using GRC. My main gripe is that downloading the reference genomic databases and GO files (if any) needs to be done by the user. But because they are all concentrated in two places (the README file points you to the correct places in NCBI and in Integr8), Warren and Setubal could have provided a script for easy downloading on relevant genomes and genome associated files from those two resources. Other than that, I like it. It’s good for the wetlab, and that is how it as originally written. But the GPL license and the simple architecture means it is good for the biohacker too: you can play with it and modify as suited to your specific needs.
And my own post-genomic moment? Like I said earlier, the term has been vacated of any initial meaning it may have had, and is here for me to cast it with new meaning. So here is my take: sitting at my laptop, running GRC on one of my favorite unannotated bugs with its close 50-odd annotated relatives (some other time on that), I realized how far we have come in our understanding of genome structures, genomes, and the ways to decipher them. Yes, you still need specialists, sophisticated software, an excellent eye for detail and ingenious experimentation to resolve the uniquness of each organism. There are many things wholesale laptop-based genome annotation can’t tell you, or worse, tell you wrong things. But hey, I can sit at home with a cheap $300 laptop, download install and run a program on it, and quickly start some serious poking at a bacterium that a few hours ago was all AGCTs. How cool and post-genomic is that?
Hallelujah.
Andrew S Warren, Joao C Setubal (2009). The Genome Reverse Compiler: an explorative annotation tool BMC Bioinformatics, 10 (1) DOI: 10.1186/1471-2105-10-35



















Your enthusiasm came through to a point that I want to find an organism, sequence it’s genome and try it this for myself!
By the way, the link to the GRC paper is broken. Small typo broke the link. 🙂
@Ricardo
Thanks for the positive feedback, Ricardo. Typo fixed.